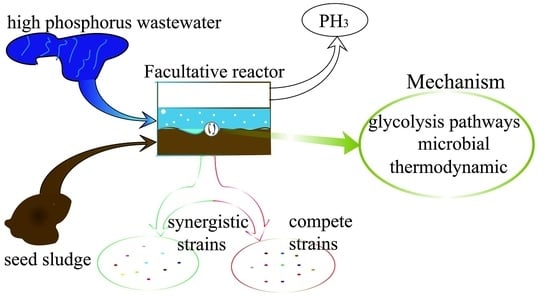
The colonies of the activated sludge samples for the three periods were columned (bar plot, combined with less than 1% of the area) at the “class” taxonomic level. As shown in
Figure 9, according to the relative abundance of strains, the effect was estimated in different periods. The relative abundance values of Bacilli in the early, middle, and best periods were 7.09%, 39.00%, and 34.55% respectively. The abundance of the Bacilli increased significantly from the initial to the mid-term by 82.8%. The abundance value of the best period decreased, which was 11.4% lower than the medium term. It is thought that the species that compete with phosphate-reducing bacteria may belong to this class; the relative abundance values of Gammaproteobacteria in the early, middle, and best periods were 0.00%, 14.97%, and 36.37% respectively. In the initial stage, the abundance value was close to zero. The best period abundance increased by 58.8% compared with the medium term. Reducing bacteria or strains that are synergistic with phosphate-reducing bacteria may belong to this class. The relative abundance values of Bacteroidia in the early, middle and best periods were 41.45%, 2.08%, and 9.43%, respectively. The mid-term abundance value decreased by 95.0% from the initial period. The best period of abundance increased by 77.9% compared with the medium term. The abundance was significantly reduced with the increase of phosphine production. It was possible to theorize that strains that compete with phosphate-reducing bacteria may belong to this class. The relative abundance values of Clostridia in the early, middle, and best periods were 30.94%, 6.36%, and 5.66%, respectively. The abundance showed a significant downward trend. The medium-term abundance value decreased by 79.4% compared with the initial period. The best period of abundance was 11.0% lower than the medium term. With the increase of phosphine production, the abundance value decreased. The strains that are competitive with phosphate-reducing bacteria may belong to the class; the relative abundance values of Actinobacteria in the early, middle, and best periods were 17.36%, 16.61%, and 1.40% respectively. The abundance value was in a downward trend. The intermediate period decreased by 4.3% compared with the initial period, and the best period was 91.6% lower than the medium term. Species that compete with phosphate-reducing bacteria may belong to this class; the relative abundance values of norank_p_Saccharibacteria in the early, middle, and best periods were 0.00%, 0.01%, and 9.65% respectively. The initial abundance was close to zero, the mid-abundance was also very small and the best period abundance improved. Analysis of phosphate-reducing bacteria or strains that are synergistic with phosphate-reducing bacteria may belong to this class; the relative abundance values of Sphingobacteria in the early, middle, and best periods were 0.00%, 7.47%, and 0.02% respectively. The relative abundance of Alphaproteobacteria in the initial, middle, and best periods were 0.01%, 6.48%, and 0.02% respectively. The relative abundance values of Flavobacteria in the early, middle, and best periods were 0.00%, 6.06%, and 0.00% respectively. Their initial abundance values were close to zero, the mid-abundance value increased and the abundance values were close to zero, which was a significant decline in the best period. The analysis suggests that the species that compete with phosphate may belong to these classes. The relative abundance values of Negativicutes in the early, middle, and best periods were 2.16%, 0.01%, and 2.49% respectively. The abundance value decreased by 99.5% in the middle of the period, which was significantly lower. The abundance value of the best period rebounded. In the medium term, its competition was at a disadvantage, resulting in a significant decrease in abundance. In the best period, due to the abundance of environmental adaptability, the competition was at a disadvantage. The rebound was not obvious. It is believed that the species that compete with phosphate may belong to this class; The relative abundance values of Others, which were less than 1%, in the initial, medium, and best periods, were 0.98%, 0.97%, and 0.40%, respectively. In summary, it can be seen that phosphate-reducing bacteria or bacteria that were synergistic with phosphate-reducing bacteria may belong to norank_p_Saccharibacteria and Gammaproteobacteria.