Fluorescence Excitation–Emission Matrix Spectroscopy and Boosting Regression Tree Model to Detect Dissolved Organic Carbon in Water
Abstract
:1. Introduction
2. Materials and Methods
2.1. Boosting Regression Tree Based Method
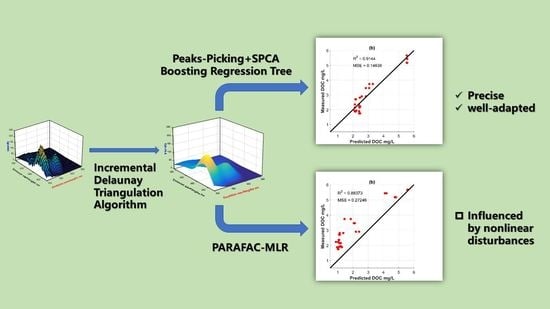
2.1.1. Incremental Delaunay Triangulation Algorithm
2.1.2. Feature Extraction
2.1.3. Boosting Regression Tree
2.2. PARAFAC-MLR Model
2.3. Assessing the Accuracy of the Model
2.4. Experiment
2.4.1. Site Descriptions and Sampling
2.4.2. Fluorescence Measurements
2.4.3. DOC Measurements
3. Results
3.1. Pre-Processing
3.2. Boosting Regression Tree Approach
3.3. PARAFAC-MLR Approach
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Williams, P.M.; Druffel, E.R.M. Radiocarbon in dissolved organic matter in the central North Pacific Ocean. Nat. Cell Biol. 1987, 330, 246–248. [Google Scholar] [CrossRef] [Green Version]
- Santinelli, C.; Ribotti, A.; Sorgente, R.; Gasparini, G.; Nannicini, L.; Vignudelli, S.; Seritti, A. Coastal dynamics and dissolved organic carbon in the western Sardinian shelf (Western Mediterranean). J. Mar. Syst. 2008, 74, 167–188. [Google Scholar] [CrossRef]
- Hansell, D.A.; Carlson, C.A. Deep-ocean gradients in the concentration of dissolved organic carbon. Nat. Cell Biol. 1998, 395, 263–266. [Google Scholar] [CrossRef]
- Avril, B. DOC dynamics in the northwestern Mediterranean Sea (DYFAMED site). Deep. Sea Res. Part II Top. Stud. Oceanogr. 2002, 49, 2163–2182. [Google Scholar] [CrossRef]
- Guo, L.; Santschi, P.H. Sedimentary sources of old high molecular weight dissolved organic carbon from the ocean margin benthic nepheloid layer. Geochim. Cosmochim. Acta 2000, 64, 651–660. [Google Scholar] [CrossRef]
- Rue, E.L.; Bruland, K.W. The role of organic complexation on ambient iron chemistry in the equatorial Pacific Ocean and the response of a mesoscale iron addition experiment. Limnol. Oceanogr. 1997, 42, 901–910. [Google Scholar] [CrossRef] [Green Version]
- Coale, K.H.; Bruland, K.W. Copper complexation in the Northeast Pacific. Limnol. Oceanogr. 1988, 33, 1084–1101. [Google Scholar] [CrossRef]
- Sharp, J.H.; Benner, R.; Bennett, L.; Carlson, C.A.; Fitzwater, S.E.; Peltzer, E.; Tupas, L.M. Analyses of dissolved organic carbon in seawater: The JGOFS EqPac methods comparison. Mar. Chem. 1995, 48, 91–108. [Google Scholar] [CrossRef]
- Sharp, J.H. Marine dissolved organic carbon: Are the older values correct? Mar. Chem. 1997, 56, 265–277. [Google Scholar] [CrossRef]
- Cook, S.; Peacock, M.; Evans, C.D.; Page, S.E.; Whelan, M.J.; Gauci, V.; Kho, L.K. Quantifying tropical peatland dissolved organic carbon (DOC) using UV-visible spectroscopy. Water Res. 2017, 115, 229–235. [Google Scholar] [CrossRef] [Green Version]
- Causse, J.; Thomas, O.; Jung, A.-V.; Thomas, M.-F. Direct DOC and nitrate determination in water using dual pathlength and second derivative UV spectro-photometry. Water Res. 2017, 108, 312–319. [Google Scholar] [CrossRef]
- Weishaar, J.L.; Aiken, G.R.; Bergamaschi, B.; Fram, M.; Fujii, R.; Mopper, K. Evaluation of Specific Ultraviolet Absorbance as an Indicator of the Chemical Composition and Reactivity of Dissolved Organic Carbon. Environ. Sci. Technol. 2003, 37, 4702–4708. [Google Scholar] [CrossRef]
- Spencer, R.G.M.; Butler, K.D.; Aiken, G.R. Dissolved organic carbon and chromophoric dissolved organic matter properties of rivers in the USA. J. Geophys. Res. Space Phys. 2012, 117, 1–14. [Google Scholar] [CrossRef]
- Peuravuori, J.; Pihlaja, K. Molecular size distribution and spectroscopic properties of aquatic humic substances. Anal. Chim. Acta 1997, 337, 133–149. [Google Scholar] [CrossRef]
- Edzwald, J.K.; Becker, W.C.; Wattier, K.L. Surrogate Parameters for Monitoring Organic Matter and THM Precursors. J. Am. Water Work. Assoc. 1985, 77, 122–132. [Google Scholar] [CrossRef]
- Carter, H.T.; Tipping, E.; Koprivnjak, J.-F.; Miller, M.; Cookson, B.; Hamilton-Taylor, J. Freshwater DOM quantity and quality from a two-component model of UV absorbance. Water Res. 2012, 46, 4532–4542. [Google Scholar] [CrossRef] [Green Version]
- Peacock, M.; Evans, C.D.; Fenner, N.; Freeman, C.; Gough, R.; Jones, T.G.; Lebron, I. UV-visible absorbance spectroscopy as a proxy for peatland dissolved organic carbon (DOC) quantity and quality: Considerations on wavelength and absorbance degradation. Environ. Sci. Process. Impacts 2014, 16, 1445–1461. [Google Scholar] [CrossRef] [Green Version]
- Downing, B.D.; Boss, E.; Bergamaschi, B.A.; Fleck, J.A.; Lionberger, M.A.; Ganju, N.K.; Schoellhamer, D.H.; Fujii, R. Quantifying fluxes and characterizing compositional changes of dissolved organic matter in aquatic systems in situ using combined acoustic and optical measurements. Limnol. Oceanogr. Methods 2009, 7, 119–131. [Google Scholar] [CrossRef] [Green Version]
- Tipping, E.; Corbishley, H.T.; Koprivnjak, J.-F.; Lapworth, D.J.; Miller, M.P.; Vincent, C.D.; Hamilton-Taylor, J. Quantification of natural DOM from UV absorption at two wavelengths. Environ. Chem. 2009, 6, 472–476. [Google Scholar] [CrossRef] [Green Version]
- Asmala, E.; Stedmon, C.; Thomas, D. Linking CDOM spectral absorption to dissolved organic carbon concentrations and loadings in boreal estuaries. Estuar. Coast. Shelf Sci. 2012, 111, 107–117. [Google Scholar] [CrossRef]
- Wang, G.-S.; Hsieh, S.-T. Monitoring natural organic matter in water with scanning spectrophotometer. Environ. Int. 2001, 26, 205–212. [Google Scholar] [CrossRef]
- Fichot, C.G.; Benner, R. A novel method to estimate DOC concentrations from CDOM absorption coefficients in coastal waters. Geophys. Res. Lett. 2011, 38. [Google Scholar] [CrossRef]
- Avagyan, A.; Runkle, B.R.K.; Kutzbach, L. Application of high-resolution spectral absorbance measurements to determine dissolved organic carbon concentration in remote areas. J. Hydrol. 2014, 517, 435–446. [Google Scholar] [CrossRef] [Green Version]
- Her, N.; Amy, G.; Foss, D.; Cho, J. Variations of Molecular Weight Estimation by HP-Size Exclusion Chromatography with UVA versus Online DOC Detection. Environ. Sci. Technol. 2002, 36, 3393–3399. [Google Scholar] [CrossRef] [PubMed]
- Korshin, G.; Benjamin, M.M.; Sletten, R.S. Adsorption of natural organic matter (NOM) on iron oxide: Effects on NOM composition and formation of organo-halide compounds during chlorination. Water Res. 1997, 31, 1643–1650. [Google Scholar] [CrossRef]
- Leenheer, J.A.; Croué, J.-P. Peer Reviewed: Characterizing Aquatic Dissolved Organic Matter. Environ. Sci. Technol. 2003, 37, 18A–26A. [Google Scholar] [CrossRef] [Green Version]
- Perminova, I.V.; Frimmel, F.H.; Kudryavtsev, A.V.; Kulikova, N.A.; Abbt-Braun, G.; Hesse, S.; Petrosyan, V.S. Molecular Weight Characteristics of Humic Substances from Different Environments as Determined by Size Exclusion Chromatography and Their Statistical Evaluation. Environ. Sci. Technol. 2003, 37, 2477–2485. [Google Scholar] [CrossRef]
- Xiao, K.; Sun, J.-Y.; Shen, Y.-X.; Liang, S.; Liang, P.; Wang, X.-M.; Huang, X. Fluorescence properties of dissolved organic matter as a function of hydrophobicity and molecular weight: Case studies from two membrane bioreactors and an oxidation ditch. Rsc Adv. 2016, 6, 24050–24059. [Google Scholar] [CrossRef]
- Belzile, C.; Roesler, C.S.; Christensen, J.P.; Shakhova, N.; Semiletov, I. Fluorescence measured using the WETStar DOM fluorometer as a proxy for dissolved matter absorption. Estuar. Coast. Shelf Sci. 2006, 67, 441–449. [Google Scholar] [CrossRef]
- Saraceno, J.F.; Pellerin, B.A.; Downing, B.D.; Boss, E.; Bachand, P.A.M.; Bergamaschi, B.A. High-frequency in situ optical measurements during a storm event: Assessing relationships between dis-solved organic matter, sediment concentrations, and hydrologic processes. J. Geophys. Res. 2009, 114. [Google Scholar] [CrossRef]
- Zhang, Y.; Liang, X.; Wang, Z.; Xu, L. A novel approach combining self-organizing map and parallel factor analysis for monitoring water quality of watersheds under non-point source pollution. Sci. Rep. 2015, 5, 16079. [Google Scholar] [CrossRef] [Green Version]
- Church, M.J.; Ducklow, H.W.; Karl, D.M. Multiyear increases in dissolved organic matter inventories at Station ALOHA in the North Pacific Subtropical Gyre. Limnol. Oceanogr. 2002, 47, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Zepp, R.G.; Sheldon, W.M.; Moran, M.A. Dissolved organic fluorophores in southeastern US coastal waters: Correction method for eliminating Rayleigh and Raman scattering peaks in excitation–emission matrices. Mar. Chem. 2004, 89, 15–36. [Google Scholar] [CrossRef]
- Soommart, S.; Paitoonwattanakij, K. Incremental Delaunay triangulation algorithm for digital terrain modelling. Thammasat Int. J. Sci. Tech. 1999, 4, 64–75. [Google Scholar]
- Sakhi, A.; Naimi, B. A hybrid refinement/decimation triangulation method for image approximation. In Proceedings of the 5th International Conference on Visual Information Engineering (VIE 2008), Xi’an, China, 29 July–1 August 2008. [Google Scholar]
- Chen, W.; Westerhoff, P.; Leenheer, J.A.; Booksh, K. Fluorescence excitation—emission matrix regional integration to quantify spectra for dissolved organic matter. Environ. Sci. Technol. 2003, 37, 5701–5710. [Google Scholar] [CrossRef]
- Friedman, J.H. Greedy function approximation: A gradient boosting machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Bro, R. PARAFAC. Tutorial and applications. Chemom. Intell. Lab. Syst. 1997, 38, 149–171. [Google Scholar] [CrossRef]
- Bro, R. Multi-Way Analysis in the Food Industry-Models, Algorithms, and Applications. 1998. Available online: https://www.semanticscholar.org/paper/MULTI-WAY-ANALYSIS-IN-THE-FOOD-INDUSTRY-Models%2C-%26-Bro/f2bdc868e33937a52c519bdf13e51a7afffbc03c#citing-papers (accessed on 6 October 2021).
- Tomasi, G.; Bro, R. PARAFAC and missing values. Chemom. Intell. Lab. Syst. 2005, 75, 163–180. [Google Scholar] [CrossRef]
- James, G.; Witten, D.; Hastie, T.; Tibshirani, R. An Introduction to Statistical Learning; Springer: New York, NY, USA, 2013; Volume 103. [Google Scholar]
- Coble, P.G. Marine optical biogeochemistry: The chemistry of ocean color. Chem. Rev. 2007, 107, 402–418. [Google Scholar] [CrossRef]
- Chen, R.; Ren, L.-F.; Shao, J.; He, Y.; Zhang, X. Changes in degrading ability, populations and metabolism of microbes in activated sludge in the treatment of phenol wastewater. RSC Adv. 2017, 7, 52841–52851. [Google Scholar] [CrossRef] [Green Version]
- Andersson, A.C.; Bro, R. The N-way Toolbox for MATLAB. Chemom. Intell. Lab. Syst. 2000, 52, 1–4. [Google Scholar] [CrossRef]
- Bro, R.; Jong, S.D. A fast non-negativity-constrained least squares algorithm. J. Chemom. 1997, 11, 393–401. [Google Scholar] [CrossRef]
- Bro, R.; Sidiropoulos, N.D. Least squares algorithms under unimodality and non-negativity constraints. J. Chemom 1998, 12, 223–247. [Google Scholar] [CrossRef]
- Hansell, D.A.; Carlson, C.A. Biogeochemistry of Marine Dissolved Organic Matter; Academic Press: Cambridge, MA, USA, 2014. [Google Scholar]
- Yang, Y.Z.; Peleato, N.M.; Legge, R.L.; Andrews, R.C. Fluorescence excitation emission matrices for rapid detection of poly-cyclic aromatic hydrocarbons and pesticides in surface waters. Environ. Sci. Water Res. Technol. 2019, 5, 315–324. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, H.; Wang, K.; Liu, Y.; Huang, P.; Yu, J.; Hou, D. Fluorescence Excitation–Emission Matrix Spectroscopy and Boosting Regression Tree Model to Detect Dissolved Organic Carbon in Water. Water 2021, 13, 3612. https://doi.org/10.3390/w13243612
Yin H, Wang K, Liu Y, Huang P, Yu J, Hou D. Fluorescence Excitation–Emission Matrix Spectroscopy and Boosting Regression Tree Model to Detect Dissolved Organic Carbon in Water. Water. 2021; 13(24):3612. https://doi.org/10.3390/w13243612
Chicago/Turabian StyleYin, Hang, Ke Wang, Yu Liu, Pingjie Huang, Jie Yu, and Dibo Hou. 2021. "Fluorescence Excitation–Emission Matrix Spectroscopy and Boosting Regression Tree Model to Detect Dissolved Organic Carbon in Water" Water 13, no. 24: 3612. https://doi.org/10.3390/w13243612