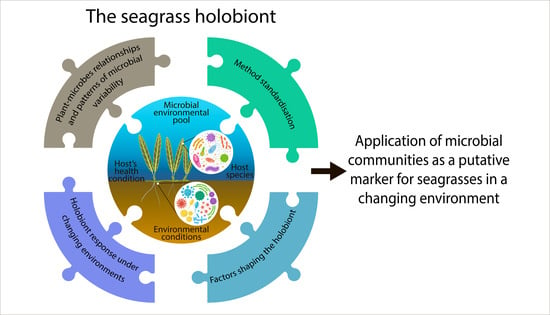
The Seagrass Holobiont: What We Know and What We Still Need to Disclose for Its Possible Use as an Ecological Indicator
Abstract
:1. Introduction
1.1. The Holobiont Concept
1.2. The Seagrass Holobiont
1.3. A Case Study: The Halophila stipulacea Holobiont
2. Hinges of the Seagrass Holobiont Composition
2.1. Plant Species
2.2. Environmental Conditions
2.3. Taxonomy and Functions
3. Key Role of Methods in Characterizing the Seagrass Holobiont (from Classical to “Omics”)
4. Mutual Influence Seagrass/Microbes: Implication and Insights for the Development of an Integrated Index
- (a)
- Occurrence and relative abundance of specific bacterial groups (taxonomy) and/or genes or metabolic pathways (functionality) can be putative markers, particularly those involved in important biogeochemical transformations within the nitrogen, sulfur and carbon cycles and thus able to affect the ecology/physiology of the seagrass host.
- (b)
- Abundance pattern and contributions of rare vs. abundant components of the holobiont microbial partner into the integrity of the seagrass host health status and the related ecological processes. For example, the rare microbial components can have greater sensitivity to both natural- and human-induced disturbances and/or exert significant impacts on seagrass ecosystem health, plasticity or resilience. Similarly, the core microbiome and the changing components of the seagrass associated microbial communities can give a different contribution to the functional structure of the microbial assemblages and thus to the ecological status of the seagrass holobiont. However, instead of a universally important seagrass–microbial core, a local or species-specific microbial core might be useful to identify.
- (c)
- Differences and roles of the microbiomes associated with the different anatomic plant compartments, which can reflect and be related to the characteristics of the environmental matrices (sediment vs. seawater). For example, the belowground compartment seems to be an extremely conservative microenvironment for seagrass associated microbial community, showing good sensitivity to the seagrass physiological condition and responding to the environmental conditions; the aboveground microbial community, instead, showed higher variability and seemed more easily influenced by the seawater conditions. Thus, the belowground microbial community may be suitable to monitor the seagrass ecological status while the aboveground microbial community may track the environmental changes, making seagrass leaves a good reference surface to monitor environmental variations or disturbances.
- (d)
- Plant surface colonization patterns/pathways and ecological interactions occurring within the seagrass epiphytic microbial community can be complex and have important implications for the holobiont health and disease (e.g., symbiosis, competition, predation, antifouling and antimicrobial activities); thus, they deserve attention and further investigation to give an insight of the host–microbial functional relationships.
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- den Hartog, C.; Kuo, J. Taxonomy and Biogeography of Seagrasses. In Seagrasses: Biology, Ecology and Conservation; Larkum, A.W.D., Orth, R.J., Duarte, C., Eds.; Springer: Dordrecht, The Netherlands, 2006; pp. 1–23. ISBN 140202942X. [Google Scholar]
- Duarte, C.M. Seagrass depth limits. Aquat. Bot. 1991, 40, 363–377. [Google Scholar] [CrossRef]
- Hemminga, M.A.; Duarte, C.M. Taxonomy and distribution. In Seagrass Ecology; Hemminga, M.A., Duarte, C.M., Eds.; Cambridge University Press: Cambridge, UK, 2000; pp. 1–26. ISBN 0-521-66184-6. [Google Scholar]
- Wright, J.P.; Jones, C.G. The concept of organisms as ecosystem engineers ten years on: Progress, limitations, and challenges. Bioscience 2006, 56, 203–209. [Google Scholar] [CrossRef] [Green Version]
- Nordlund, L.M.; Koch, E.W.; Barbier, E.B.; Creed, J.C. Seagrass Ecosystem Services and Their Variability across Genera and Geographical Regions. PLoS ONE 2016, 11, e0163091. [Google Scholar] [CrossRef] [Green Version]
- Beck, M.W.; Heck, K.L.; Able, K.W.; Childers, D.L.; Eggleston, D.B.; Gillanders, B.M.; Halpern, B.; Hays, C.G.; Hoshino, K.; Minello, T.J.; et al. The identification, conservation, and management of estuarine and marine nurseries for fish and invertebrates. Bioscience 2001, 51, 633–641. [Google Scholar] [CrossRef]
- Jiang, Z.; Cui, L.; Liu, S.; Zhao, C.; Wu, Y.; Chen, Q.; Yu, S.; Li, J.; He, J.; Fang, Y.; et al. Historical changes in seagrass beds in a rapidly urbanizing area of Guangdong Province: Implications for conservation and management. Glob. Ecol. Conserv. 2020, 22, e01035. [Google Scholar] [CrossRef]
- Jeyabaskaran, R.; Jayasankar, J.; Ambrose, T.V.; Valsalan, K.C.V.; Divya, N.D.; Raji, N.; Vysakhan, P.; John, S.; Prema, D.; Kaladharan, P.; et al. Conservation of seagrass beds with special reference to associated species and fishery resources. J. Mar. Biol. Assoc. India 2018, 60, 62–70. [Google Scholar] [CrossRef]
- Duarte, C.M.; Marbà, N.; Gacia, E.; Fourqurean, J.W.; Beggins, J.; Barrón, C.; Apostolaki, E.T. Seagrass community metabolism: Assessing the carbon sink capacity of seagrass meadows. Glob. Biogeochem. Cycles 2010, 24, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Duarte, C.M.; Middelburg, J.J.; Caraco, N. Major role of marine vegetation on the oceanic carbon cycle. Biogeosciences 2005, 1, 173–180. [Google Scholar] [CrossRef] [Green Version]
- Orth, R.J.; Carruthers, T.J.B.; Dennison, W.C.; Duarte, C.M.; Fourqurean, J.W.; Kenneth, L.H.J.; Hughes, A.R.; Kendrick, G.A.; Kenworthy, W.J.; Olyarnik, S.; et al. A Global Crisis for Seagrass Ecosystems. Bioscience 2006, 56, 987–996. [Google Scholar] [CrossRef] [Green Version]
- Waycott, M.; Duarte, C.M.; Carruthers, T.J.B.; Orth, R.J.; Dennison, W.C.; Olyarnik, S.; Calladine, A.; Fourqurean, J.W.; Heck, K.L.; Hughes, A.R.; et al. Accelerating loss of seagrasses across the globe threatens coastal ecosystems. Proc. Natl. Acad. Sci. USA 2009, 106, 12377–12381. [Google Scholar] [CrossRef] [Green Version]
- Short, F.T.; Neckles, H.A. The effects of global climate change on seagrasses. Aquat. Bot. 1999, 63, 169–196. [Google Scholar] [CrossRef]
- Duarte, B.; Martins, I.; Rosa, R.; Matos, A.R.; Roleda, M.Y.; Reusch, T.B.H.; Engelen, A.H.; Serrão, E.A.; Pearson, G.A.; Marques, J.C.; et al. Climate Change Impacts on Seagrass Meadows and Macroalgal Forests: An Integrative Perspective on Acclimation and Adaptation Potential. Front. Mar. Sci. 2018, 5. [Google Scholar] [CrossRef] [Green Version]
- de los Santos, C.B.; Krause-Jensen, D.; Alcoverro, T.; Marbà, N.; Duarte, C.M.; Van Katwijk, M.M.; Pérez, M.; Romero, J.; Sánchez-lizaso, J.L.; Trayter, G.M.; et al. Recent trend reversal for declining European seagrass meadows. Nat. Commun. 2019, 10, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pergent, G.; Pergent-Martini, C.; Boudouresque, C.F. Utilisation de l’herbier a Posidonia oceanica comme indicateur biologique de la qualité du milieu littoral en méditerranée: État des connaissances. Mésogée 1995, 54, 3–27. [Google Scholar]
- Martínez-Crego, B.; Vergés, A.; Alcoverro, T.; Romero, J. Selection of multiple seagrass indicators for environmental biomonitoring. Mar. Ecol. Prog. Ser. 2008, 361, 93–109. [Google Scholar] [CrossRef]
- Roca, G.; Alcoverro, T.; Krause-Jensen, D.; Balsby, T.J.S.; Van Katwijk, M.M.; Marbà, N.; Santos, R.; Arthur, R.; Mascaró, O.; Fernández-Torquemada, Y.; et al. Response of seagrass indicators to shifts in environmental stressors: A global review and management synthesis. Ecol. Indic. 2016, 63, 310–323. [Google Scholar] [CrossRef] [Green Version]
- Rotini, A.; Anello, L.; Di Bernardo, M.; Giallongo, A.; Valiante, L.; Migliore, L. Comparative analysis of bed density, total phenol content and protein expression pattern in Posidonia oceanica (L.) Delile. Open J. Ecol. 2013, 3, 438–444. [Google Scholar] [CrossRef] [Green Version]
- Migliore, L.; Rotini, A.; Randazzo, D.; Albanese, N.N.; Giallongo, A. Phenols content and 2-D electrophoresis protein pattern: A promising tool to monitor Posidonia meadows health state. BMC Ecol. 2007, 7, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Martínez, M.; López-López, A.; Calleja, M.L.; Marbà, N.; Duarte, C.M. Bacterial community dynamics in a seagrass (Posidonia oceanica) meadow sediment. Estuaries Coasts 2008, 32, 276–286. [Google Scholar] [CrossRef] [Green Version]
- Marco-Noales, E.; Ordax, M.; Armando, D.; Lopez, M.M.; Saavedra, M.J.; Martinez-Murcia, A.; Garcias, N.; Marbá, N.; Duarte, C.M. Microbiota associated with P. oceanica in Western Mediterranean Sea. Mod. Multidiscip. Appl. Microbiol. 2006, 342–350. [Google Scholar] [CrossRef]
- Hassenrück, C.; Hofmann, L.C.; Bischof, K.; Ramette, A. Seagrass biofilm communities at a naturally CO2-rich vent. Environ. Microbiol. Rep. 2015, 7, 516–525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mejia, A.Y.; Rotini, A.; Lacasella, F.; Bookman, R.; Thaller, M.C.; Shem-tov, R.; Winters, G.; Migliore, L. Assessing the ecological status of seagrasses using morphology, biochemical descriptors and microbial community analyses. A study in Halophila stipulacea (Forsk.) Aschers meadows in the. Ecol. Indic. 2016, 60, 1150–1163. [Google Scholar] [CrossRef]
- Rotini, A.; Mejia, A.Y.; Costa, R.; Migliore, L.; Winters, G. Ecophysiological Plasticity and Bacteriome Shift in the Seagrass Halophila stipulacea along a Depth Gradient in the Northern Red Sea. Front. Plant Sci. 2017, 7, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, B.C.; Bougoure, J.; Ryan, M.H.; Bennett, W.W.; Colmer, T.D.; Joyce, N.K.; Olsen, Y.S.; Kendrick, G.A. Oxygen loss from seagrass roots coincides with colonisation of sulphide-oxidising cable bacteria and reduces sulphide stress. ISME J. 2018, 13, 707–719. [Google Scholar] [CrossRef] [Green Version]
- Crump, B.C.; Wojahn, J.M.; Tomas, F.; Mueller, R.S. Metatranscriptomics and amplicon sequencing reveal mutualisms in seagrass microbiomes. Front. Microbiol. 2018, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Cúcio, C.; Overmars, L.; Engelen, A.H.; Muyzer, G. Metagenomic Analysis Shows the Presence of Bacteria Related to Free-Living Forms of Sulfur-Oxidizing Chemolithoautotrophic Symbionts in the Rhizosphere of the Seagrass Zostera marina. Front. Mar. Sci. 2018, 5, 1–15. [Google Scholar] [CrossRef]
- Martin, B.C.; Gleeson, D.; Statton, J.; Siebers, A.R.; Grierson, P.; Ryan, M.H.; Kendrick, G.A. Low light availability alters root exudation and reduces putative beneficial microorganisms in seagrass roots. Front. Microbiol. 2018, 8, 1–16. [Google Scholar] [CrossRef]
- Tarquinio, F.; Jeremy, B.; Koenders, A.; Hyndes, G.A. Microorganisms facilitate uptake of dissolved organic nitrogen by seagrass leaves. ISME J. 2018, 12, 2796–2800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rotini, A.; Conte, C.; Seveso, D.; Montano, S.; Galli, P.; Vai, M.; Migliore, L.; Mejia, A. Daily variation of the associated microbial community and the Hsp60 expression in the Maldivian seagrass Thalassia hemprichii. J. Sea Res. 2020, 156, 101835. [Google Scholar] [CrossRef]
- Martin, B.C.; Sanchez Alcaron, M.; Gleeson, D.; Middleton, J.A. Root microbiomes as indicators of seagrass health. FEMS Microbiol. Ecol. 2020, 96, fiz201. [Google Scholar] [CrossRef] [PubMed]
- Zilber-Rosenberg, I.; Rosenberg, E. Role of microorganisms in the evolution of animals and plants: The hologenome theory of evolution. FEMS Microbiol. Rev. 2008, 32, 723–735. [Google Scholar] [CrossRef]
- Valdez, S.R.; Zhang, Y.S.; van der Heide, T.; Vanderklift, M.A.; Tarquinio, F.; Orth, R.J.; Silliman, B.R. Positive Ecological Interactions and the Success of Seagrass Restoration. Front. Mar. Sci. 2020, 7, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Uku, J.; Björk, M.; Bergman, B.; Díez, B. Characterization and comparison of prokaryotic epiphytes associated with three East African seagrasses. J. Phycol. 2007, 43, 768–779. [Google Scholar] [CrossRef]
- Crump, B.C.; Koch, E.W. Attached Bacterial Populations Shared by Four Species of Aquatic Angiosperms. Appl. Environ. Microbiol. 2008, 74, 5948–5957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wahbeh, M.I.; Mahasneh, A.M. Heterotrophic Bacteria Attached To Leaves, Rhizomes And Roots of Three Seagrass Species From Aqaba (Jordan). Aquat. Bot. 1984, 20, 87–96. [Google Scholar] [CrossRef]
- Cúcio, C.; Engelen, A.H.; Costa, R.; Muyzer, G.; Costa, R. Rhizosphere Microbiomes of European Seagrasses Are Selected by the Plant, But Are Not Species Specific. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Ugarelli, K.; Laas, P.; Stingl, U. The Microbial Communities of Leaves and Roots Associated with Turtle Grass (Thalassia testudinum) and Manatee Grass (Syringodium filliforme) are Distinct from Seawater and Sediment Communities, but Are Similar between Species and Sampling Sites. Microorganisms 2018, 7, 4. [Google Scholar] [CrossRef] [Green Version]
- Trevathan-Tackett, S.M.; Sherman, C.D.H.; Huggett, M.J.; Campbell, A.H.; Laverock, B.; Hurtado-McCormick, V.; Seymour, J.R.; Firl, A.; Messer, L.F.; Ainsworth, T.D.; et al. A horizon scan of priorities for coastal marine microbiome research. Nat. Ecol. Evol. 2019, 3, 1509–1520. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, L.G.E.; Leray, M.; O’Dea, A.; Yuen, B.; Peixoto, R.S.; Pereira, T.J.; Bik, H.M.; Coil, D.A.; Duffy, J.E.; Herre, E.A.; et al. Host-associated microbiomes drive structure and function of marine ecosystems. PLoS Biol. 2019, 17, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dittami, S.; Arboleda, E.; Auguet, J.-C.; Bigalke, A.; Briand, E.; Cardenas, P.; Cardini, U.; Decelle, J.; Engelen, A.; Eveillard, D.; et al. A community perspective on the concept of marine holobionts: Current status, challenges, and future directions. PeerJ 2019, 1–22. [Google Scholar] [CrossRef]
- Margulis, L.; Fester, R. Symbiosis as a Source of Evolutionary Innovation: Special Ion and Morphogenesis; MIT Press: Cambridge, UK, 1991; ISBN 0-262-13269-9. [Google Scholar]
- O’Malley, M.A. Endosymbiosis and its implications for evolutionary theory. Proc. Natl. Acad. Sci. USA 2015, 112, 10270–10277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gray, M.W. Lynn Margulis and the endosymbiont hypothesis: 50 years later. Mol. Biol. Cell 2017, 28, 1285–1287. [Google Scholar] [CrossRef] [PubMed]
- Baedke, J.; Fábregas-Tejeda, A.; Nieves Delgado, A. The holobiont concept before Margulis. J. Exp. Zool. Part B Mol. Dev. Evol. 2020, 334, 149–155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenberg, E.; Zilber-Rosenberg, I. Microbes Drive Evolution of Animals and Plants: The Hologenome Concept. MBio 2016, 3, 193–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roughgarden, J.; Gilbert, S.F.; Rosenberg, E.; Zilber-Rosenberg, I.; Lloyd, E.A. Holobionts as Units of Selection and a Model of Their Population Dynamics and Evolution. Biol. Theory 2018, 13, 44–65. [Google Scholar] [CrossRef]
- Bordenstein, S.R.; Theis, K.R. Host biology in light of the microbiome: Ten principles of holobionts and hologenomes. PLoS Biol. 2015, 13, 1–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Bustos Muñoz, M.T.; Guirao, J.L.G.; Vera López, J.A.; Campuzano, A.V. The hologenome concept of evolution after 10 years. Qual. Theory Dyn. Syst. 2018, 14, 265–280. [Google Scholar] [CrossRef]
- Theis, K.R.; Dheilly, N.M.; Klassen, J.L.; Brucker, R.M.; Baines, J.F.; Lloyd, E.A.; Sapp, J.; Vandenkoornhuyse, P.; Zilber-Rosenberg, I.; Rosenberg, E.; et al. Getting the Hologenome Concept Right: An Eco-Evolutionary Framework for Hosts and Their Microbiomes. Msystems 2016, 1, e00028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bang, C.; Dagan, T.; Deines, P.; Dubilier, N.; Duschl, W.J.; Fraune, S.; Hentschel, U.; Hirt, H.; Hülter, N.; Lachnit, T.; et al. Metaorganisms in extreme environments: Do microbes play a role in organismal adaptation? Zoology 2018, 127, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Simon, J.C.; Marchesi, J.R.; Mougel, C.; Selosse, M.A. Host-microbiota interactions: From holobiont theory to analysis. Microbiome 2019, 7, 1–5. [Google Scholar] [CrossRef]
- Vandenkoornhuyse, P.; Quaiser, A.; Duhamel, M.; Le Van, A.; Dufresne, A. The importance of the microbiome of the plant holobiont. New Phytol. 2015, 206, 1196–1206. [Google Scholar] [CrossRef]
- Sullivan, B.K.; Trevathan-Tackett, S.M.; Neuhauser, S.; Govers, L.L. Review: Host-pathogen dynamics of seagrass diseases under future global change. Mar. Pollut. Bull. 2018, 134, 75–88. [Google Scholar] [CrossRef] [Green Version]
- Pitlik, S.D.; Koren, O. How holobionts get sick-toward a unifying scheme of disease. Microbiome 2017, 5. [Google Scholar] [CrossRef] [PubMed]
- Longford, S.R.; Campbell, A.H.; Nielsen, S.; Case, R.J.; Kjelleberg, S.; Steinberg, P.D. Interactions within the microbiome alter microbial interactions with host chemical defences and affect disease in a marine holobiont. Sci. Rep. 2019, 9, 1363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Egan, S.; Gardiner, M. Microbial dysbiosis: Rethinking disease in marine ecosystems. Front. Microbiol. 2016, 7, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ursell, L.K.; Metcalf, J.M.; Parfrey, L.W.; Knight, R. Defining the Human Microbiome. Nat. Rev. Microbiol. 2012, 70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, B.; Yao, M.; Lv, L.; Ling, Z.; Li, L. The Human Microbiota in Health and Disease. Engineering 2017, 3, 71–82. [Google Scholar] [CrossRef]
- Lerner, A.; Neidhöfer, S.; Matthias, T. The Gut Microbiome Feelings of the Brain: A Perspective for Non-Microbiologists. Microorganisms 2017, 5, 66. [Google Scholar] [CrossRef]
- Carthey, A.J.R.; Blumstein, D.T.; Gallagher, R.V.; Tetu, S.G.; Gillings, M.R. Conserving the holobiont. Funct. Ecol. 2019, 34, 764–776. [Google Scholar] [CrossRef]
- Thompson, J.R.; Rivera, H.E.; Closek, C.J.; Medina, M. Microbes in the coral holobiont: Partners through evolution, development, and ecological interactions. Front. Cell. Infect. Microbiol. 2015, 4, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Reshef, L.; Koren, O.; Loya, Y.; Zilber-Rosenberg, I.; Rosenberg, E. The Coral Probiotic Hypothesis. Environ. Microbiol. 2006, 8, 2068–2073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pita, L.; Rix, L.; Slaby, B.M.; Franke, A.; Hentschel, U. The sponge holobiont in a changing ocean: From microbes to ecosystems. Microbiome 2018, 6, 46. [Google Scholar] [CrossRef] [PubMed]
- Vollmer, S.V.; Baker, A.C.; Coffroth, M.-A.; Harvell, C.D.; Medina, M. Understanding the coral holobiont through science and scuba. Smithson. Contrib. Mar. Sci. 2013, 39, 173–186. [Google Scholar]
- Hassani, M.A.; Durán, P.; Hacquard, S. Microbial interactions within the plant holobiont. Encycl. Syst. Biol. 2018, 6, 902. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Cañizares, C.; Jorrín, B.; Poole, P.S.; Tkacz, A. Understanding the holobiont: The interdependence of plants and their microbiome. Curr. Opin. Microbiol. 2017, 38, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Lemanceau, P.; Barret, M.; Mazurier, S.; Mondy, S.; Pivato, B.; Fort, T.; Vacher, C. Plant Communication with Associated Microbiota in the Spermosphere, Rhizosphere and Phyllosphere. In How Plants Communicate with Their Biotic Environment; Becard, G., Ed.; Academic Press: Cambridge, MA, USA, 2017; Volume 82, pp. 101–133. ISBN 9780128014318. [Google Scholar]
- Long, S.R. Rhizobium-legume nodulation: Life together in the underground. Cell 1989, 56, 203–214. [Google Scholar] [CrossRef]
- Masson-Boivin, C.; Sachs, J.L. Symbiotic nitrogen fixation by rhizobia—The roots of a success story. Curr. Opin. Plant Biol. 2018, 44, 7–15. [Google Scholar] [CrossRef] [Green Version]
- van der Loos, L.M.; Eriksson, B.K.; Falcão Salles, J. The Macroalgal Holobiont in a Changing Sea. Trends Microbiol. 2019, 27, 635–650. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.P.; Reddy, C.R.K. Seaweed-microbial interactions: Key functions of seaweed-associated bacteria. FEMS Microbiol. Ecol. 2014, 88, 213–230. [Google Scholar] [CrossRef]
- Egan, S.; Harder, T.; Burke, C.; Steinberg, P.; Kjelleberg, S.; Thomas, T. The seaweed holobiont: Understanding seaweed-bacteria interactions. FEMS Microbiol. Rev. 2013, 37, 462–476. [Google Scholar] [CrossRef] [Green Version]
- Thurber, R.V.; Willner-Hall, D.; Rodriguez-Mueller, B.; Desnues, C.; Edwards, R.A.; Angly, F.; Dinsdale, E.; Kelly, L.; Rohwer, F. Metagenomic analysis of stressed coral holobionts. Environ. Microbiol. 2009, 11, 2148–2163. [Google Scholar] [CrossRef] [PubMed]
- Marzinelli, E.M.; Campbell, A.H.; Valdes, Z.E.; Vergés, A.; Nielsen, S.; Wernberg, T.; de Bettignies, T.; Bennett, S.; Caporaso, J.G.; Thomas, T.; et al. Continental-scale variation in seaweed host-associated bacterial communities is a function of host condition, not geography. Environ. Microbiol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Minich, J.J.; Morris, M.M.; Brown, M.; Doane, M.; Edwards, M.S.; Michael, T.P.; Dinsdale, E.A. Elevated temperature drives kelp microbiome dysbiosis, while elevated carbon dioxide induces water microbiome disruption. PLoS ONE 2018, 13, e0192772. [Google Scholar] [CrossRef] [PubMed]
- Ugarelli, K.; Chakrabarti, S.; Laas, P.; Stingl, U. The Seagrass Holobiont and Its Microbiome. Microorganisms 2017, 5, 81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tarquinio, F.; Hyndes, G.A.; Laverock, B.; Koenders, A.; Säwström, C. The seagrass holobiont: Understanding seagrass-bacteria interactions and their role in seagrass ecosystem functioning. FEMS Microbiol. Lett. 2019, 366, fnz057. [Google Scholar] [CrossRef]
- Seymour, J.R.; Laverock, B.; Nielsen, D.A.; Macreadie, P.I. The Microbiology of Seagrass. In Seagrasses of Australia; Larkum, A.W.D., Kendrick, G.A., Ralph, P.J., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2018; pp. 343–392. ISBN 978-3-319-71352-6. [Google Scholar] [CrossRef]
- Garcias-Bonet, N.; Arrieta, J.M.; de Santana, C.N.; Duarte, C.M.; Marbà, N. Endophytic bacterial community of a Mediterranean marine angiosperm (Posidonia oceanica). Front. Microbiol. 2012, 3, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Croft, M.T.; Lawrence, A.D.; Raux-Deery, E.; Warren, M.J.; Smith, A.G. Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature 2005, 438, 90–93. [Google Scholar] [CrossRef]
- Touchette, B.W.; Burkholder, J.A.M. Review of nitrogen and phosphorus metabolism in seagrasses. J. Exp. Mar. Bio. Ecol. 2000, 250, 133–167. [Google Scholar] [CrossRef]
- Brodersen, K.E.; Koren, K.; Moßhammer, M.; Ralph, P.J.; Ku, M.; Santner, J. Seagrass-Mediated Phosphorus and Iron Solubilization in Tropical Sediments. Environ. Sci. Technol. 2017, 51, 14155–14163. [Google Scholar] [CrossRef] [PubMed]
- Holmer, M.; Andersen, F.O.; Nielsen, S.L.; Boschker, H.T.S. The importance of mineralization based on sulfate reduction for nutrient regeneration in tropical seagrass sediments. Aquat. Bot. 2001, 71, 1–17. [Google Scholar] [CrossRef]
- Welsh, D.T. Nitrogen fixation in seagrass meadows: Regulation, plant-bacteria interactions and significance to primary productivity. Ecol. Lett. 2000, 3, 58–71. [Google Scholar] [CrossRef]
- Zimmerman, R.; Smith, R.; Alberte, R. Is growth of eelgrass nitrogen limited? A numerical simulation of the effects of light and nitrogen on the growth dynamics of Zostera marina. Mar. Ecol. Prog. Ser. 1987, 41, 167–176. [Google Scholar] [CrossRef]
- Celdrán, D.; Espinosa, E.; Sánchez-Amat, A.; Marín, A. Effects of epibiotic bacteria on leaf growth and epiphytes of the seagrass Posidonia oceanica. Mar. Ecol. Prog. Ser. 2012, 456, 21–27. [Google Scholar] [CrossRef] [Green Version]
- Brodersen, K.E.; Siboni, N.; Nielsen, D.A.; Pernice, M.; Ralph, P.J.; Seymour, J.; Kuhl, M. Seagrass rhizosphere microenvironment alters plant-associated microbial community composition. Environ. Microbiol. 2018, 20, 2854–2864. [Google Scholar] [CrossRef] [PubMed]
- Brodersen, K.E.; Michael, K.; Nielsen, D.A.; Pedersen, O.; Larkum, A.W.D. Rhizome, Root/Sediment Interactions, Aerenchyma and Internal Pressure Changes in Seagrasses. In Seagrasses of Australia; Larkum, A.W.D., Kendrick, G.A., Ralph, P.J., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2018; pp. 393–418. ISBN 9783319713540. [Google Scholar]
- Nielsen, L.B.; Finster, K.; Welsh, D.T.; Donelly, A.; Herbert, R.A.; De Wit, R.; Lomstein, B.A. Sulphate reduction and nitrogen fixation rates associated with roots, rhizomes and sediments from Zostera noltii and Spartina maritima meadows. Environ. Microbiol. 2001, 3, 63–71. [Google Scholar] [CrossRef]
- Lehnen, N.; Marchant, H.K.; Schwedt, A.; Milucka, J.; Lott, C.; Weber, M.; Dekaezemacker, J.; Seah, B.K.B.; Hach, P.F.; Mohr, W.; et al. High rates of microbial dinitrogen fixation and sulfate reduction associated with the Mediterranean seagrass Posidonia oceanica. Syst. Appl. Microbiol. 2016, 39, 476–483. [Google Scholar] [CrossRef] [PubMed]
- Hurtado-McCormick, V.; Kahlke, T.; Petrou, K.; Jeffries, T.; Ralph, P.J.; Seymour, J.R.; Hill, R.T. Regional and Microenvironmental Scale Characterization of the Zostera muelleri Seagrass Microbiome. Front. Microbiol. 2019, 10, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Carbonero, F.; Oakley, B.B.; Purdy, K.J. Metabolic Flexibility as a Major Predictor of Spatial Distribution in Microbial Communities. PLoS ONE 2014, 9, e85105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, F.; Zhang, X.; Zhang, Q.; Liu, F.; Zhang, J.; Gong, J. Seagrass (Zostera marina) colonization promotes the accumulation of diazotrophic bacteria and alters the relative abundances of specific bacterial lineages involved in benthic carbon and sulfur cycling. Appl. Environ. Microbiol. 2015, 81, 6901–6914. [Google Scholar] [CrossRef] [Green Version]
- Ettinger, C.L.; Voerman, S.E.; Lang, J.M.; Stachowicz, J.J.; Eisen, J.A. Microbial communities in sediment from Zostera marina patches, but not the Z. marina leaf or root microbiomes, vary in relation to distance from patch edge. PeerJ 2017, 2017, 1–25. [Google Scholar] [CrossRef] [Green Version]
- Ettinger, C.L.; Williams, S.L.; Abbott, J.M.; Stachowicz, J.J.; Eisen, J.A. Microbiome succession during ammonification in eelgrass bed sediments. PeerJ 2017, 2017, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Garcias-Bonet, N.; Arrieta, J.M.; Duarte, C.M.; Marbà, N. Nitrogen-fixing bacteria in Mediterranean seagrass (Posidonia oceanica) roots. Aquat. Bot. 2016, 131, 57–60. [Google Scholar] [CrossRef] [Green Version]
- Jensen, S.I.; Kühl, M.; Priemé, A. Different bacterial communities associated with the roots and bulk sediment of the seagrass Zostera marina. FEMS Microbiol. Ecol. 2007, 62. [Google Scholar] [CrossRef] [Green Version]
- Kristensen, E.; Holmer, M. Decomposition of plant materials in marine sediment exposed to different electron acceptors (O2, NO3, and SO42−), with emphasis on substrate origin, degradation kinetics, and the role of bioturbation. Geochim. Cosmochim. Acta 2001, 65, 419–433. [Google Scholar] [CrossRef]
- Apostolaki, E.T.; Holmer, M.; Santinelli, V.; Karakassis, I. Species-specific response to sulfide intrusion in native and exotic Mediterranean seagrasses under stress. Mar. Environ. Res. 2018, 134, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Hasler-Sheetal, H.; Fragner, L.; Holmer, M.; Weckwerth, W. Diurnal effects of anoxia on the metabolome of the seagrass Zostera marina. Metabolomics 2015, 11, 1208–1218. [Google Scholar] [CrossRef] [Green Version]
- Brodersen, K.E.; Lichtenberg, M.; Paz, L.C.; Kühl, M. Epiphyte-cover on seagrass (Zostera marina L.) leaves impedes plant performance and radial O2 loss from the below-ground tissue. Front. Mar. Sci. 2015, 2, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Van Der Heide, T.; Govers, L.L.; De Fouw, J.; Olff, H.; Van Der Geest, M.; Van Katwijk, M.M.; Piersma, T.; Van De Koppel, J.; Silliman, B.R.; Smolders, A.J.P.; et al. A three-stage symbiosis forms the foundation of seagrass ecosystems. Science 2012, 336, 1432–1434. [Google Scholar] [CrossRef] [Green Version]
- Pereg, L.L.; Lipkin, Y.; Sar, N. Different niches of the Halophila stipulacea seagrass bed harbor distinct populations of nitrogen fixing bacteria. Mar. Biol. 1994, 119, 327–333. [Google Scholar] [CrossRef]
- Garcias-Bonet, N.; Fusi, M.; Ali, M.; Shaw, D.R.; Saikaly, P.E.; Daffonchio, D.; Duarte, C.M. High denitrification and anaerobic ammonium oxidation contributes to net nitrogen loss in a seagrass ecosystem in the central Red Sea. Biogeosciences 2018, 15, 7333–7346. [Google Scholar] [CrossRef] [Green Version]
- Hamisi, M.; Díez, B.; Lyimo, T.; Ininbergs, K.; Bergman, B. Epiphytic cyanobacteria of the seagrass Cymodocea rotundata: Diversity, diel nifH expression and nitrogenase activity. Environ. Microbiol. 2013, 5. [Google Scholar] [CrossRef]
- Mayavu, P.; Sugesh, S.; Ravindran, V.J. Antibacterial activity of seagrass species against biofilm forming bacteria. Res. J. Microbiol. 2009, 4, 314–319. [Google Scholar] [CrossRef] [Green Version]
- Howayada, H.A.E.-H.; Eman, R.H.; Abeer, N.S. Molecular Identification, Antimicrobial and Antioxidant Activities of the Tropical Seagrass Halophila stipulacea Grown in El-Bardawil Lake. Aust. J. Basic Appl. Sci. 2012, 6, 474–481. [Google Scholar]
- Babuselvam, M.; Farook, K.A.; Abideen, S.; Peer Mohamed, M.; Uthiraselvam, M. Screening of Antibacterial Activity of Mangrove Leaf. Int. J. Appl. Microbiol. Sci. 2012, 13, 20–25. [Google Scholar]
- Satheesh, S.; Ba-Akdah, M.A.; Al-Sofyani, A.A. Natural antifouling compound production by microbes associated with marine macroorganisms—A review. Electron. J. Biotechnol. 2016, 21, 26–35. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Chen, W.; Wang, G.; Dai, S.; Zhou, D.; Zhao, H.; Guo, Y.; Ouyang, Y.; Li, X. Culture-dependent diversity of Actinobacteria associated with seagrass (Thalassia hemprichii). Afr. J. Microbiol. Res. 2012, 6, 87–94. [Google Scholar] [CrossRef]
- Armstrong, E.; Yan, L.; Boyd, K.G.; Wright, P.C.; Burgess, J.G. The symbiotic role of marine microbes on living surfaces. Hydrobiologia 2001, 461, 37–40. [Google Scholar] [CrossRef]
- Rao, D.; Webb, J.S.; Kjelleberg, S. Competitive Interactions in Mixed-Species Biofilms Containing the Marine Bacterium Pseudoalteromonas tunicata Dhana. Appl. Environ. Microbiol. 2005, 71, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Ravikumar, S.; Gnanadesigan, M.; Saravanan, A.; Monisha, N.; Brindha, V.; Muthumari, S. Antagonistic properties of seagrass associated Streptomyces sp. RAUACT-1: A source for anthraquinone rich compound. Asian Pac. J. Trop. Med. 2012, 5, 887–890. [Google Scholar] [CrossRef]
- Ravikumar, S.; Thajuddin, N.; Suganthi, P.; Jacob Lnbaneson, S.; Vinodkumar, T. Bioactive potential of seagrass bacteria against human bacterial pathogens. J. Environ. Biol. 2010, 31, 387–389. [Google Scholar] [PubMed]
- Marhaeni, B.; Radjasa, O.K.; Bengen, D.G.; Kaswadji, R.F. Screening of bacterial symbionts of seagrass Enhalus sp. against biofilm-forming bacteria. J. Coast. Dev. 2010, 13, 126–132. [Google Scholar]
- Marhaeni, B.; Radjasa, O.K.; Sabdono, A.; Sudoyo, H. Antifouling Activity of Bacterial Symbionts of Seagrasses against Marine Biofilm-Forming Bacteria. J. Environ. Prot. (Irvine Calif.) 2011, 2, 1245–1249. [Google Scholar] [CrossRef] [Green Version]
- Inaba, N.; Trainer, V.L.; Onishi, Y.; Ishii, K.I.; Wyllie-Echeverria, S.; Imai, I. Algicidal and growth-inhibiting bacteria associated with seagrass and macroalgae beds in Puget Sound, WA, USA. Harmful Algae 2017, 62, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Fahimipour, A.K.; Kardish, M.R.; Lang, J.M.; Green, J.L.; Eisen, J.A.; Stachowicz, J.J. Globalscale structure of the eelgrass microbiome. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kohn, T.; Rast, P.; Kallscheuer, N.; Wiegand, S.; Boedeker, C.; Jetten, M.S.M.; Jeske, O.; Vollmers, J.; Kaster, A.K.; Rohde, M.; et al. The Microbiome of Posidonia oceanica Seagrass Leaves Can Be Dominated by Planctomycetes. Front. Microbiol. 2020, 11, 1458. [Google Scholar] [CrossRef] [PubMed]
- García-Martínez, M.; Kuo, J.; Kilminster, K.; Walker, D.; Rosselló-Mora, R.; Duarte, C.M. Microbial colonization in the seagrass Posidonia spp. roots. Mar. Biol. Res. 2005, 1, 388–395. [Google Scholar] [CrossRef] [Green Version]
- Fraser, M.W.; Statton, J.; Hovey, R.K.; Laverock, B.; Kendrick, G.A. Seagrass derived organic matter influences biogeochemistry, microbial communities, and seedling biomass partitioning in seagrass sediments. Plant Soil 2016, 400, 133–146. [Google Scholar] [CrossRef]
- Fourqurean, J.W.; Duarte, C.M.; Kennedy, H.; Marbà, N.; Holmer, M.; Mateo, M.A.; Apostolaki, E.T.; Kendrick, G.A.; Krause-Jensen, D.; McGlathery, K.J.; et al. Seagrass ecosystems as a globally significant carbon stock. Nat. Geosci. 2012, 5, 505–509. [Google Scholar] [CrossRef]
- Liu, S.; Jiang, Z.; Zhou, C.; Wu, Y.; Arbi, I.; Zhang, J.; Huang, X.; Trevathan-Tacket, S.M. Leaching of dissolved organic matter from seagrass leaf litter and its biogeochemical implications. Acta Oceanol. Sin. 2018, 37, 84–90. [Google Scholar] [CrossRef]
- Macreadie, P.I.; Atwood, T.B.; Seymour, J.R.; Fontes, M.L.S.; Sanderman, J.; Nielsen, D.A.; Connolly, R.M. Vulnerability of seagrass blue carbon to microbial attack following exposure to warming and oxygen. Sci. Total Environ. 2019, 686, 264–275. [Google Scholar] [CrossRef]
- Dang, H. Grand Challenges in Microbe-Driven Marine Carbon Cycling Research. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Bouillon, S.; Boschker, H.T.S.; Brussel, V.U. Bacterial carbon sources in coastal sediments: A cross-system analysis based on stable isotope data of biomarkers. Biogeosciences 2006, 3, 175–185. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Jiang, Z.; Wu, Y.; Zhang, J.; Arbi, I.; Ye, F.; Huang, X.; Macreadie, P.I. Effects of nutrient load on microbial activities within a seagrass-dominated ecosystem: Implications of changes in seagrass blue carbon. Mar. Pollut. Bull. 2017, 117, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Trevathan-Tackett, S.M.; Jeffries, T.C.; Macreadie, P.I.; Manojlovic, B.; Ralph, P. Long-term decomposition captures key steps in microbial breakdown of seagrass litter. Sci. Total Environ. 2020, 705, 135806. [Google Scholar] [CrossRef] [PubMed]
- Trevathan-tackett, S.M.; Seymour, J.R.; Nielsen, D.A.; Macreadie, P.I.; Jeffries, T.C.; Sanderman, J.; Baldock, J.; Howes, J.M.; Steven, A.D.L.; Ralph, P.J. Sediment anoxia limits microbial-driven seagrass carbon remineralization under warming conditions. FEMS Microbiol. Ecol. 2017, 93, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Pedersen, M.Ø.; Serrano, O.; Mateo, M.Á.; Holmer, M. Temperature effects on decomposition of a Posidonia oceanica mat. Aquat. Microb. Ecol. 2011, 65, 169–182. [Google Scholar] [CrossRef] [Green Version]
- Trevathan-Tackett, S.M.; Macreadie, P.I.; Sanderman, J.; Baldock, J.; Howes, J.M.; Ralph, P.J. A Global Assessment of the Chemical Recalcitrance of Seagrass Tissues: Implications for Long-Term Carbon Sequestration. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Macreadie, P.I.; Hardy, S.S.S. Response of seagrass “Blue Carbon” stocks to increased water temperatures. Diversity 2018, 10, 115. [Google Scholar] [CrossRef] [Green Version]
- Winters, G.; Beer, S.; Willette, D.A.; Viana, I.G.; Chiquillo, K.L. The Tropical Seagrass Halophila stipulacea: Reviewing What We Know from Its Native and Invasive Habitats, Alongside Identifying Knowledge Gaps. Front. Mar. Sci. 2020, 7. [Google Scholar] [CrossRef]
- Gambi, M.C.; Barbieri, F.; Bianchi, C.N. New record of the alien seagrass Halophila stipulacea (Hydrocharitaceae) in the western Mediterranean: A further clue to changing Mediterranean Sea biogeography. Mar. Biodivers. Rec. 2009, 2. [Google Scholar] [CrossRef]
- Sghaier, Y.R.; Zakhama-Sraieb, R.; Mouelhi, S.; Vazquez, M.; Valle, C.; Ramos-Espla, A.A.; Astier, J.M.; Verlaque, M.; Charfi-Cheikhrouha, F. Review of alien marine macrophytes in Tunisia. Mediterr. Mar. Sci. 2016, 17, 109–123. [Google Scholar] [CrossRef] [Green Version]
- Sghaier, Y.R.; Zakhama-Sraieb, R.; Benamer, I.; Charfi-Cheikhrouha, F. Occurrence of the seagrass Halophila stipulacea (Hydrocharitaceae) in the southern Mediterranean Sea. Bot. Mar. 2011, 54, 575–582. [Google Scholar] [CrossRef]
- Willette, D.A.; Ambrose, R.F. Effects of the invasive seagrass Halophila stipulacea on the native seagrass, Syringodium filiforme, and associated fish and epibiota communities in the Eastern Caribbean. Aquat. Bot. 2012, 103, 74–82. [Google Scholar] [CrossRef]
- Vera, B.; Collado-Vides, L.; Moreno, C.; van Tussenbroek, B.I. Halophila stipulacea (Hydrocharitaceae): A Recent Introduction to the Continental Waters of Venezuela. Caribb. J. Sci. 2014, 48, 66–70. [Google Scholar] [CrossRef]
- Scheibling, R.E.; Patriquin, D.G.; Filbee-Dexter, K. Distribution and abundance of the invasive seagrass Halophila stipulacea and associated benthic macrofauna in Carriacou, Grenadines, Eastern Caribbean. Aquat. Bot. 2018, 144, 1–8. [Google Scholar] [CrossRef]
- Willette, D.A.; Chalifour, J.; Debrot, A.O.D.; Engel, M.S.; Miller, J.; Oxenford, H.A.; Short, F.T.; Steiner, S.C.C.; Védie, F. Continued expansion of the trans-Atlantic invasive marine angiosperm Halophila stipulacea in the eastern Caribbean. Aquat. Bot. 2014, 112, 98–102. [Google Scholar] [CrossRef]
- van Tussenbroek, B.I.; Govers, L.L.; van der Heide, T.; van Katwijk, M.M.; Bouma, T.J.; Leuven, R.S.E.W. Non-native seagrass Halophila stipulacea forms dense mats under eutrophic conditions in the Caribbean. J. Sea Res. 2016, 115, 1–5. [Google Scholar] [CrossRef]
- Smulders, F.O.H.; Vonk, J.A.; Engel, M.S.; Christianen, M.J.A. Expansion and fragment settlement of the non-native seagrass Halophila stipulacea in a Caribbean bay. Mar. Biol. Res. ISSN 2017, 13, 967–974. [Google Scholar] [CrossRef]
- Streftaris, N.; Zenetos, A. Alien marine species in the Mediterranean—The 100 “worst invasives” and their impact. Mediterr. Mar. Sci. 2006, 7, 87–118. [Google Scholar] [CrossRef] [Green Version]
- Bianchi, C.N.; Morri, C. Global sea warming and “tropicalization” of the Mediterranean Sea: Biogeographic and ecological aspects. Biogeogr. J. Integr. Biogeogr. 2003, 24. [Google Scholar] [CrossRef] [Green Version]
- Sghier, Y.R.; Zakhama-Sraieb, R.; Charfi-Cheikhrouha, F. Effects of the invasive seagrass Halophila stipulacea on the native seagrass Cymodocea Nodosa. In Proceedings of the 5th Mediterranean Symposium on Marine Vegetation, Portorož, Slovenia, 27–28 October 2014; pp. 27–28. [Google Scholar]
- Sharon, Y.; Silva, J.; Santos, R.; Runcie, J.W.; Chernihovsky, M.; Beer, S. Photosynthetic responses of Halophila stipulacea to a light gradient. II. Acclimations following transplantation. Aquat. Biol. 2009, 7, 153–157. [Google Scholar] [CrossRef]
- Sharon, Y.; Beer, S. Diurnal movements of chloroplasts in Halophila stipulacea and their effect on PAM fluorometric measurements of photosynthetic rates. Aquat. Bot. 2008, 88, 273–276. [Google Scholar] [CrossRef]
- Oscar, M.A.; Barak, S.; Winters, G. The Tropical Invasive Seagrass, Halophila stipulacea, Has a Superior Ability to Tolerate Dynamic Changes in Salinity Levels Compared to Its Freshwater Relative, Vallisneria americana. Front. Plant Sci. 2018, 9, 1–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexandre, A.; Luis, J.; Georgiou, D.; Alexandre, A.; Luis, J.; Santos, R. Temperature is not a limiting factor for the expansion of Halophila stipulacea throughout the whole Mediterranean Sea. Mar. Ecol. Prog. Ser. 2016, 544, 159–167. [Google Scholar] [CrossRef]
- Azcárate-García, T.; Beca-Carretero, P.; Villamayor, B.; Stengel, D.B.; Winters, G. Responses of the seagrass Halophila stipulacea to depth and spatial gradients in its native region (Red Sea): Morphology, in situ growth and biomass production. Aquat. Bot. 2020, 165, 103252. [Google Scholar] [CrossRef]
- Beca-Carretero, P.; Rotini, A.; Mejia, A.; Migliore, L.; Vizzini, S.; Winters, G. Halophila stipulacea descriptors in the native area (Red Sea): A baseline for future comparisons with native and non-native populations. Mar. Environ. Res. 2019, 153, 104828. [Google Scholar] [CrossRef] [PubMed]
- Procaccini, G.; Acunto, S.; Famà, P.; Maltagliati, F. Structural, morphological and genetic variability in Halophila stipulacea (Hydrocharitaceae) populations in the western Mediterranean. Mar. Biol. 1999, 135, 181–189. [Google Scholar] [CrossRef]
- Nguyen, H.M.; Yadav, N.S.; Barak, S.; Lima, F.P.; Sapir, Y.; Winters, G. Responses of Invasive and Native Populations of the Seagrass Halophila stipulacea to Simulated Climate Change. Front. Mar. Sci. 2020, 6, 1–16. [Google Scholar] [CrossRef]
- Sharon, Y.; Levitan, O.; Spungin, D.; Berman-Frank, I.; Beer, S. Photoacclimation of the seagrass Halophila stipulacea to the dim irradiance at its 48-m depth limit. Limnol. Oceanogr. 2011, 56, 357–362. [Google Scholar] [CrossRef]
- Schwarz, A.M.; Hellblom, F. The photosynthetic light response of Halophila stipulacea growing along a depth gradient in the Gulf of Aqaba, the Red Sea. Aquat. Bot. 2002, 74, 263–272. [Google Scholar] [CrossRef]
- Manh Nguyen, H.; Savva, I.; Kleitou, P.; Kletou, D.; Lima, F.P.; Sapir, Y.; Winters, G. Seasonal dynamics of native and invasive Halophila stipulacea populations—A case study from the northern Gulf of Aqaba and the eastern Mediterranean Sea. Aquat. Bot. 2020, 162, 103205. [Google Scholar] [CrossRef] [Green Version]
- Hulings, N.C. The Ecology, Biometry and Biomass of the Seagrass Halophila stipulacea Along the Jordanian Coast of the Gulf of Aqaba. Bot. Mar. 1979, 22, 425–430. [Google Scholar] [CrossRef]
- Lipkin, Y. Quantitative Aspects of Seagrass Communities, Particularly of Those Dominated by Halophila Stipulacea, in Sinai (Northern Red Sea). Aquat. Bot. 1979, 7, 119–128. [Google Scholar] [CrossRef]
- Weidner, S.; Arnold, W.; Pühler, A. Diversity of uncultured microorganisms associated with the seagrass Halophila stipulacea estimated by restriction fragment length polymorphism analysis of PCR-amplified 16s rRNA genes. Appl. Environ. Microbiol. 1996, 62, 766–771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weidner, S.; Arnold, W.; Stackebrandt, E.; Pühler, A. Phylogenetic analysis of bacterial communities associated with leaves of the seagrass Halophila stipulacea by a culture-independent small-subunit rRNA gene approach. Microb. Ecol. 2000, 39, 22–31. [Google Scholar] [CrossRef]
- Dang, H.; Li, T.; Chen, M.; Huang, G. Cross-ocean distribution of Rhodobacterales bacteria as primary surface colonizers in temperate coastal marine waters. Appl. Environ. Microbiol. 2008, 74, 52–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elifantz, H.; Horn, G.; Ayon, M.; Cohen, Y.; Minz, D. Rhodobacteraceae are the key members of the microbial community of the initial biofilm formed in Eastern Mediterranean coastal seawater. FEMS Microbiol. Ecol. 2013, 85, 348–357. [Google Scholar] [CrossRef] [Green Version]
- Buchan, A.; González, J.M.; Moran, M.A. Overview of the marine Roseobacter lineage. Appl. Environ. Microbiol. 2005, 71, 5665–5677. [Google Scholar] [CrossRef] [Green Version]
- Campbell, A.M.; Fleisher, J.; Sinigalliano, C.; White, J.R.; Lopez, J.V. Dynamics of marine bacterial community diversity of the coastal waters of the reefs, inlets, and wastewater outfalls of southeast Florida. Microbiol. Open. 2015, 4, 390–408. [Google Scholar] [CrossRef] [PubMed]
- Lemanceau, P.; Blouin, M.; Muller, D.; Moënne-Loccoz, Y. Let the Core Microbiota Be Functional. Trends Plant Sci. 2017, 22, 583–595. [Google Scholar] [CrossRef]
- Garrity, G.M.; Bell, J.A.; Lilburn, T. Pseudomonadaceae. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2015; ISBN 9781118960608. [Google Scholar]
- Bitam, F.; Ciavatta, M.L.; Carbone, M.; Manzo, E.; Mollo, E.; Gavagnin, M. Chemical analysis of fl avonoid constituents of the seagrass Halophila stipulacea: First finding of malonylated derivatives in marine phanerogams. Biochem. Syst. Ecol. 2010, 38, 686–690. [Google Scholar] [CrossRef]
- Zidorn, C. Secondary metabolites of seagrasses (Alismatales and Potamogetonales; Alismatidae): Chemical diversity, bioactivity, and ecological function. Phytochemistry 2016, 124, 5–28. [Google Scholar] [CrossRef] [PubMed]
- Defranoux, F.; Mollo, E. Molecular Interactions as Drivers of Changes in Marine Ecosystems. In Co-Evolution of Secondary Metabolites; Mérillon, J.-M., Ramawat, K.G., Eds.; Springer Nature Switzerland AG: Cham, Switzerland, 2020; pp. 121–133. ISBN 9783319768878. [Google Scholar]
- Mierziak, J.; Kostyn, K.; Kulma, A. Flavonoids as important molecules of plant interactions with the environment. Molecules 2014, 19, 16240–16265. [Google Scholar] [CrossRef] [PubMed]
- Mannino, A.M.; Micheli, C. Ecological function of phenolic compounds from mediterranean fucoid algae and seagrasses: An overview on the genus Cystoseira sensu lato and Posidonia oceanica (L.) Delile. J. Mar. Sci. Eng. 2020, 8, 19. [Google Scholar] [CrossRef] [Green Version]
- Gordon-Bradley, N.; Lymperopoulou, D.S.; Williams, H.N. Differences in bacterial community structure on Hydrilla verticillata and Vallisneria Americana in a freshwater spring. Microbes Environ. 2014, 29, 67–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcias-Bonet, N.; Eguíluz, V.; Díaz-Rúa, R.; Duarte, C. Host-association as major driver of microbiome structure and composition in Red Sea seagrass ecosystems. Environ. Microbiol. 2020, 22. [Google Scholar] [CrossRef]
- Webb, S.J.; Rabsatt, T.; Erazo, N.; Bowman, J.S. Impacts of Zostera eelgrasses on microbial community structure in San Diego coastal waters. Elem. Sci. Anthr. 2019, 7. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Ling, J.; Dong, J.; Chen, B.; Zhang, Y.; Zhang, Y.; Wang, Y. Illumina-based analysis the microbial diversity associated with Thalassia hemprichii in Xincun Bay, South China Sea. Ecotoxicology 2015, 24, 1548–1556. [Google Scholar] [CrossRef]
- Bengtsson, M.M.; Bühler, A.; Brauer, A.; Dahlke, S.; Schubert, H. Eelgrass Leaf Surface Microbiomes Are Locally Variable and Highly Correlated with Epibiotic Eukaryotes. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Conte, C.; Rotini, A.; Winters, G.; Vasquez, M.I.; Piazza, G.; Kletou, D.; Migliore, L. Elective affinities or random choice within the seagrass holobiont? The case of the native Posidonia oceanica (L.) Delile and the invasive Halophila stipulacea (Forssk.) Asch. from the same site (Limassol, Cyprus). Aquat. Bot. under review.
- Ling, J.; Lin, X.; Zhang, Y.; Zhou, W.; Yang, Q.; Lin, L.; Zeng, S.; Zhang, Y.; Wang, C.; Ahmad, M.; et al. Community composition and transcriptional activity of ammonia-oxidizing prokaryotes of seagrass Thalassia hemprichii in coral reef ecosystems. Front. Microbiol. 2018, 9, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mansson, M.; Gram, L.; Larsen, T.O. Production of bioactive secondary metabolites by marine Vibrionaceae. Mar. Drugs 2011, 9, 1440–1468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ismail, A.; Ktari, L.; Ahmed, M.; Bolhuis, H.; Bouhaouala-Zahar, B.; Stal, L.J.; Boudabbous, A.; El Bour, M. Heterotrophic bacteria associated with the green alga Ulva rigida: Identification and antimicrobial potential. J. Appl. Phycol. 2018, 30, 2883–2899. [Google Scholar] [CrossRef]
- Campbell, A.H.; Marzinelli, E.M.; Gelber, J.; Steinberg, P.D. Spatial variability of microbial assemblages associated with a dominant habitat-forming seaweed. Front. Microbiol. 2015, 6, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Burke, C.; Steinberg, P.; Rusch, D.; Kjelleberg, S.; Thomas, T. Bacterial community assembly based on functional genes rather than species. Proc. Natl. Acad. Sci. USA 2011, 108, 14288–14293. [Google Scholar] [CrossRef] [Green Version]
- Mishra, A.K.; Mohanraju, R. Epiphytic Bacterial Communities in Seagrass Meadows of Oligotrophic Waters of Andaman Sea. Open Access Libr. J. 2018, 5. [Google Scholar] [CrossRef]
- Marzinelli, E.M.; Qiu, Z.; Dafforn, K.A.; Johnston, E.L.; Steinberg, P.D.; Mayer-Pinto, M. Coastal urbanisation affects microbial communities on a dominant marine holobiont. NPJ Biofilms Microbiomes 2018, 4. [Google Scholar] [CrossRef] [PubMed]
- Devereux, R.; Yates, D.F.; Aukamp, J.; Quarles, R.L.; Jordan, S.J.; Stanley, R.S.; Eldridge, P.M. Interactions of Thalassia testudinum and sediment biogeochemistry in Santa Rosa sound, NW Florida. Mar. Biol. Res. 2011, 7, 317–331. [Google Scholar] [CrossRef]
- Bourque, A.S.; Vega-Thurber, R.; Fourqurean, J.W. Microbial community structure and dynamics in restored subtropical seagrass sediments. Aquat. Microb. Ecol. 2015, 74, 43–57. [Google Scholar] [CrossRef] [Green Version]
- Donnelly, A.P.; Herbert, R.A. Bacterial interactions in the rhizosphere of seagrass communities in shallow coastal lagoons. J. Appl. Microbiol. 1998, 85, 151S–160S. [Google Scholar] [CrossRef]
- Fraser, M.W.; Gleeson, D.B.; Grierson, P.F.; Laverock, B.; Kendrick, G.A. Metagenomic Evidence of Microbial Community Responsiveness to Phosphorus and Salinity Gradients in Seagrass Sediments. Front. Microbiol. 2018, 9, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Song, Z.; Zhang, H.; Liu, P.; Hu, X. Seagrass vegetation affect the vertical organization of microbial communities in sediment. Mar. Environ. Res. 2020, 162, 105174. [Google Scholar] [CrossRef] [PubMed]
- Gribben, P.E.; Nielsen, S.; Seymour, J.R.; Bradley, D.J.; West, M.N.; Thomas, T. Microbial communities in marine sediments modify success of an invasive macrophyte. Sci. Rep. 2017, 7, 9845. [Google Scholar] [CrossRef] [Green Version]
- Lamb, J.B.; Van De Water, J.A.J.M.; Bourne, D.G.; Altier, C.; Hein, M.Y.; Fiorenza, E.A.; Abu, N.; Jompa, J.; Harvell, C.D. Seagrass ecosystems reduce exposure to bacterial pathogens of humans, fishes, and invertebrates. Science 2017, 355, 731–733. [Google Scholar] [CrossRef] [PubMed]
- Roth-Schulze, A.J.; Zozaya-Valdés, E.; Steinberg, P.D.; Thomas, T. Partitioning of functional and taxonomic diversity in surface-associated microbial communities. Environ. Microbiol. 2016, 18, 4391–4402. [Google Scholar] [CrossRef] [PubMed]
- Sale, P.F. Coexistence of coral reef fishes—A lottery for living space. Environ. Biol. Fishes Vol. 1978, 3, 85–102. [Google Scholar] [CrossRef]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; Mcdonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Rebecca, L.; Thurber, V.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Malmer, D.; Langille, M.G.I.; Way, S.F.; Knight, R. Which is more important for classifying microbial communities: Who’s there or what they can do? ISME J. 2014, 8, 2357–2359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, J.H.; Park, S.E.; Kang, S.J.; Oh, T.K. Rheinheimera aquimaris sp. nov., isolated from seawater of the East Sea in Korea. Int. J. Syst. Evol. Microbiol. 2007, 57, 1386–1390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nielsen, J.; Liesack, W.; Finster, K. Desulfovibrio zosterae sp. nov., a new sulfate reducer isolated from surface-sterilized roots of the seagrass Zostera marina. Int. J. Syst. Bacteriol. 1999, 49, 859–865. [Google Scholar] [CrossRef] [PubMed]
- Cifuentes, A.; Antón, J.; Benlloch, S.; Donnelly, A.; Herbert, R.A.; Rodríguez-Valera, F. Prokaryotic diversity in Zostera noltii-colonized marine sediments. Appl. Environ. Microbiol. 2000, 66, 1715–1719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alsaffar, Z.; Pearman, J.K.; Cúrdia, J.; Ellis, J.; Calleja, M.L.; Ruiz-Compean, P.; Roth, F.; Villalobos, R.; Jones, B.H.; Morán, X.A.G.; et al. The role of seagrass vegetation and local environmental conditions in shaping benthic bacterial and macroinvertebrate communities in a tropical coastal lagoon. Sci. Rep. 2020, 10, 13550. [Google Scholar] [CrossRef] [PubMed]
- Kuo, J.; McComb, A.; Cambridge, M.L. Ultrastructure of the seagrass rhizosphere. New. Phytol. 1981, 89, 139–143. [Google Scholar] [CrossRef]
- Novak, R. A study in ultra-ecology: Microorganisms of the seagrass Posidonia oceanica (L.) Delile. Mar. Ecol. 1984, 5, 143–190. [Google Scholar] [CrossRef]
- Vohník, M.; Borovec, O.; Kolařík, M. Communities of Cultivable Root Mycobionts of the Seagrass Posidonia oceanica in the Northwest Mediterranean Sea Are Dominated by a Hitherto Undescribed Pleosporalean Dark Septate Endophyte. Microb. Ecol. 2016, 71, 442–451. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.; Wang, C.; Zhang, X.; Gong, J. Community Structure and Abundance of Archaea in a Zostera marina Meadow: A Comparison between Seagrass-Colonized and Bare Sediment Sites. Archaea 2019, 2019, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Ettinger, C.L.; Eisen, J.A. Characterization of the Mycobiome of the Seagrass, Zostera marina, Reveals Putative Associations with Marine Chytrids. Front. Microbiol. 2019, 10, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Wagner, A.O.; Praeg, N.; Reitschuler, C.; Illmer, P. Effect of DNA extraction procedure, repeated extraction and ethidium monoazide (EMA)/propidium monoazide (PMA) treatment on overall DNA yield and impact on microbial fingerprints for bacteria, fungi and archaea in a reference soil. Appl. Soil Ecol. 2015, 93, 56–64. [Google Scholar] [CrossRef]
- Jones, M.B.; Highlander, S.K.; Anderson, E.L.; Li, W.; Dayrit, M.; Klitgord, N.; Fabani, M.M.; Seguritan, V.; Green, J.; Pride, D.T.; et al. Library preparation methodology can influence genomic and functional predictions in human microbiome research. Proc. Natl. Acad. Sci. USA 2015, 112, 14024–14029. [Google Scholar] [CrossRef] [Green Version]
- Escobar-Zepeda, A.; De León, A.V.P.; Sanchez-Flores, A. The road to metagenomics: From microbiology to DNA sequencing technologies and bioinformatics. Front. Genet. 2015, 61, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jünemann, S.; Sedlazeck, F.J.; Prior, K.; Albersmeier, A.; John, U.; Kalinowski, J.; Mellmann, A.; Goesmann, A.; von Haeseler, A.; Stoye, J.; et al. Updating benchtop sequencing performance comparison. Nat. Biotechnol. 2013, 31, 294–296. [Google Scholar] [CrossRef] [Green Version]
- Quail, M.A.; Smith, M.; Coupland, P.; Otto, T.D.; Harris, S.R.; Connor, T.R.; Bertoni, A.; Swerdlow, H.P.; Gu, Y. A tale of three next generation sequencing platforms: Comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genom. 2012, 13, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing MOTHUR: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Asnicar, F.; Bai, Y.B.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotech. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Nilakanta, H.; Drews, K.L.; Firrell, S.; Foulkes, M.A.; Jablonski, K.A. A review of software for analyzing molecular sequences. BMC Res. Notes 2014, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailén, M.; Bressa, C.; Larrosa, M.; González-Soltero, R. Bioinformatic strategies to address limitations of 16rRNA short-read amplicons from different sequencing platforms. J. Microbiol. Methods 2020, 169. [Google Scholar] [CrossRef] [PubMed]
- Balvočiute, M.; Huson, D.H. SILVA, RDP, Greengenes, NCBI and OTT—How do these taxonomies compare? BMC Genom. 2017, 18, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Yilmaz, P.; Parfrey, L.W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glöckner, F.O. The SILVA and “all-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2014, 42, 643–648. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Federhen, S. The NCBI Taxonomy database. Nucleic Acids Res. 2012, 40, 136–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; Desantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.S.; Spakowicz, D.J.; Hong, B.Y.; Petersen, L.M.; Demkowicz, P.; Chen, L.; Leopold, S.R.; Hanson, B.M.; Agresta, H.O.; Gerstein, M.; et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franssen, S.U.; Gu, J.; Bergmann, N.; Winters, G.; Klostermeier, U.C.; Rosenstiel, P.; Bornberg-Bauer, E. Transcriptomic resilience to global warming in the seagrass Zostera marina, a marine foundation species. Proc. Natl. Acad. Sci. USA 2011, 108, 19276–19281. [Google Scholar] [CrossRef] [Green Version]
- Davey, P.A.; Pernice, M.; Ashworth, J.; Kuzhiumparambil, U.; Szabó, M.; Dolferus, R.; Ralph, P.J. A new mechanistic understanding of light-limitation in the seagrass Zostera muelleri. Mar. Environ. Res. 2018, 134, 55–67. [Google Scholar] [CrossRef] [PubMed]
- Caruso, G. Microbes and their use as Indicators of Pollution. J. Pollut. Eff. Control 2013, 1, 1–2. [Google Scholar] [CrossRef] [Green Version]
- Cordier, T.; Lanzén, A.; Apothéloz-Perret-Gentil, L.; Stoeck, T.; Pawlowski, J. Embracing Environmental Genomics and Machine Learning for Routine Biomonitoring. Trends Microbiol. 2019, 27, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Borja, A.; Franco, J.; Pérez, V. A marine Biotic Index to establish the ecological quality of soft-bottom benthos within European estuarine and coastal environments. Mar. Pollut. Bull. 2000, 40, 1100–1114. [Google Scholar] [CrossRef]
- Borja, A. Testing the efficiency of a bacterial community-based index (microgAMBI) to assess distinct impact sources in six locations around the world. Ecol. Indic. 2018, 85, 594–602. [Google Scholar] [CrossRef]
- Muxika, I.; Borja, Á.; Bonne, W. The suitability of the marine biotic index (AMBI) to new impact sources along European coasts. Ecol. Indic. 2005, 5, 19–31. [Google Scholar] [CrossRef]
- Aires, T.; Serrão, E.A.; Engelen, A.H. Host and environmental specificity in bacterial communities associated to two highly invasive marine species (genus Asparagopsis). Front. Microbiol. 2016, 7, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Serebryakova, A.; Aires, T.; Viard, F.; Serrao, E.A.; Engelen, A.H. Summer shifts of bacterial communities associated with the invasive brown seaweed Sargassum muticum are location and tissue dependent. PLoS ONE 2018, 13, e0206734. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Song, Q.; Li, Q.; Zhang, H.; Luo, X.; Zheng, Z. Damage of heavy metals to Vallisneria natans (V. natans) and characterization of microbial community in biofilm. Aquat. Toxicol. 2020, 225. [Google Scholar] [CrossRef] [PubMed]

| Seagrass Species | Seagrass Descriptors and Environmental Parameters | Plant Part and/or Environmental Matrix a | Study Area | Methods and Target Organisms (in Brackets) | Main Results | References |
|---|---|---|---|---|---|---|
| Amphibolis antarctica, Halodule uninervis | Phosphorus and salinity gradients | Sediment | Shark Bay, WA, Australia | Amplicon sequencing of 16S rRNA genes. Hydrolytic enzymes measurement with standard colorimetric methods (β-glucosidase assayed with 4-nitrophenyl β-glucopyranoside, acid phosphatase, and alkaline phosphatase assayed with 4-nitrophenyl phosphate disodium hexahydrate) (Bacteria) | Adaptations within the microbial community point to potential coevolution between seagrasses and sediment microorganisms may help both groups thrive in the extreme environment | Fraser et al., 2018 [190] |
| Cymodocea serrulata, Halodule uninervis, Halophila ovalis | Different light treatments | Rhizosphere | Useless Loop in Shark Bay, Western Australia | Amplicon sequencing of the 16S rRNA genes (Bacteria) | The effect of light reduction on root exudation and the consequent reduction of putative beneficial microorganisms | Martin et al., 2018 [29] |
| Enhalus acroides | CO2-vents proximity | Phyllosphere | Papua New Guinea | Quantitative technique ARISA and Amplicon sequencing of 16S rRNA and 18S rRNA gene (Bacteria, Eukaryotes) | Bacterial community showed a site-related pattern at the CO2 vents with an increase of bacterial types associated with coral diseases | Hassenrück et al., 2015 [23] |
| Halophila ovalis | Healthy vs. stressed meadows: biomass, productivity and F-sulphide | Rhizosphere | Multiple locations across South-west Western Australia | Amplicon sequencing of the 16S rRNA (Bacteria) | Suggested the use of the epiphytic microbes to detected clues of stress that other seagrass health metrics failed to detect | Martin et al., 2020 [32] |
| Halophila ovalis, Zostera muelleri | Radial oxygen loss around root tips | Rhizosphere | Swan River estuary, Western Australia | Confocal fluorescence in situ hybridization, oxygen, sulphide microsensor, Amplicon sequencing of the 16S rRNA genes (Bacteria) | Roots leaking oxygen had a higher abundance of sulphide-oxidizing cable bacteria | Martin et al., 2018 [26] |
| Halophila stipulacea | Morphometry, total phenols, photosynthetic pigments. Anthropogenic pressure, granulometry, TOC% | Phyllosphere, rhizosphere | Gulf of Aqaba, Israel | Amplicon sequencing of 16S rRNA genes (Bacteria) | Proposed assessing the ecological status of seagrasses using their microbiota | Mejia et al., 2016 [24] |
| Halophila stipulacea | Morphometry, total phenols, photosynthetic pigments. Depth gradient, granulometry, TOC% | Phyllosphere, rhizosphere | Gulf of Aqaba, Israel | Amplicon sequencing of 16S rRNA genes (Bacteria) | High variability was observed in the epiphytic microbiota along a depth gradient | Rotini et al., 2017 [25] |
| Posidonia oceanica | Mortality rate | Phyllosphere, rhizosphere | Western Mediterranean Sea | 16S rRNA gene clone library (Bacteria) | Pseudoalteromonas was more abundant in the meadow with the highest mortality rate | Marco-Noales et al., 2006 [22] |
| Posidonia oceanica | Regression rate | Rhizosphere | Balearic Island, Spain | 16S rRNA gene clone library, DAPI, FISH (Bacteria) | Suggested a link between the documented regression of the meadow and the decline of the rhizosphere microbial community | García-Martinez et al., 2008 [21] |
| Thalassia hemprichii | Three different reef system Ammonium, nitrate, nitrite, and phosphate in seawater | Phyllosphere, rhizosphere, bulk sediment | Sanya Bay and Yongxing Island, South China Sea | DNA and cDNA qPCR targeting amoA genes [ammonia monooxygenase-subunit] (Bacteria) | Significant relationships between the abundance of archaeal amoA gene in rhizosphere and the ammonium concentration in sediments and nitrogen content in plant tissue | Ling et al., 2018 [180] |
| Thalassia hemprichii | Hsp60, Hsp70 expression in leaves, morphometry in different day time | Phyllosphere, rhizosphere | Lagoon of Magoodhoo island, Maldives | Amplicon sequencing of 16S rRNA gene (Bacteria) | Coupled daily changes of microbiome composition and HSP60 expression. Microbiota variation is an early clue of change of seagrass physiological state | Rotini et al., 2020 [31] |
| Vallisneria natans | Heavy metals exposure (Cu2+, Pb2+, Cd2+ and mixture). Plant growth, water chemistry and heavy metals bioaccumulation | Phyllosphere | Tiancun Horticultural Company, Shanghai, China | High-throughput amplicon sequencing of the 16S rRNA gene (Bacteria) | Abundances and structures of biofilm communities changed in response to heavy metal stress exposure | Huang et al., 2020 [232] |
| Zostera spp. | Presence/absence of seagrass, inside vs. outside the bay | Seawater column | San Diego Bay, USA | Amplicon sequencing 16S rRNA gene sequencing, flow cytometry (Bacteria) | The current flow rather than the seagrass presence shapes the seawater column microbial community | Webb et al., 2019 [176] |
| Zostera japonica, Zostera marina | Mixed meadows, different sites | Phyllosphere, Rhizosphere, seawater | Netarts Bay, OR, United States | Amplicon sequencing of the 16S rRNA genes and metatranscriptomics (Bacteria) | Microbe–plant mutualisms support the health and growth of aquatic plants | Crump et al., 2018 [27] |
| Zostera japonica, Zostera marina | Vegetated vs. degraded areas | Rhizosphere | Swan Lake, China | High-throughput amplicon sequencing of the 16S rRNA gene (Bacteria) | Different microbiota found in vegetated and degraded areas. Vibrionaceae could lead to seagrass degradation | Sun et al., 2020 [191] |
| Zostera marina | Leaf area. Depth, lagoon/open coast | Phyllosphere | In- and out-side lagoons on the German Baltic Sea coast | Amplicon sequencing of 16S and 18S rRNA genes (Bacteria, Eukaryotes) | Correlation between eukaryotic diversity and prokaryotic microbiome | Bengtsson et al., 2017 [178] |
| Zostera marina | Inner/edge plants position in the meadow | Phyllosphere, rhizosphere, sediment | Bodega Bay, USA | Amplicon sequencing of 16S rRNA genes (Bacteria) | Seagrass microbiota do not feel the edge effect | Ettinger et al., 2017 [96] |
| Zostera marina | Presence/absence of seagrass, distance from the shore and along the reefs | Seawater column | Spermonde Archipelago, Indonesia | Amplicon sequencing 16S rRNA gene sequencing and Enterococcus assays. (Bacteria) | Seagrass presence caused 50% reduction in the relative abundance of potential bacterial pathogens | Lamb et al., 2017 [193] |
| Zostera muelleri | Radial O2 loss and H2S concentration at the basal leaf meristem (Microsensor) | Rhizosphere | Narra-Been Lagoon, NSW, Australia | Amplicon sequencing of the 16S rRNA genes (Bacteria) | Seagrass-mediated alterations of rhizosphere geochemistry result in shifts of the rhizosphere microbial community composition | Brodersen et al., 2018 [89] |
| Zostera muelleri | Plant sections | Upper leaf, lower leaf, sheath, roots and rhizomes | New South Wales, Australia | Amplicon sequencing of 16S and 18S rRNA gene, ITS2 (5.8S) rRNA gene (Bacteria, Eukaryotes, Fungi) | Identified a “core” microbiome likely play key roles in seagrass physiology, ecology and biogeochemistry | Hurtado-McCormick et al., 2019 [93] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Conte, C.; Rotini, A.; Manfra, L.; D’Andrea, M.M.; Winters, G.; Migliore, L. The Seagrass Holobiont: What We Know and What We Still Need to Disclose for Its Possible Use as an Ecological Indicator. Water 2021, 13, 406. https://doi.org/10.3390/w13040406
Conte C, Rotini A, Manfra L, D’Andrea MM, Winters G, Migliore L. The Seagrass Holobiont: What We Know and What We Still Need to Disclose for Its Possible Use as an Ecological Indicator. Water. 2021; 13(4):406. https://doi.org/10.3390/w13040406
Chicago/Turabian StyleConte, Chiara, Alice Rotini, Loredana Manfra, Marco Maria D’Andrea, Gidon Winters, and Luciana Migliore. 2021. "The Seagrass Holobiont: What We Know and What We Still Need to Disclose for Its Possible Use as an Ecological Indicator" Water 13, no. 4: 406. https://doi.org/10.3390/w13040406