Characterization of the Spatial Variation of Microbial Communities in a Decentralized Subtropical Wastewater Treatment Plant Using Passive Methods
Abstract
:1. Introduction
2. Materials and Methods
2.1. Site Description
2.2. Water Quality Analyses
2.3. Dna Extraction and High-Throughput Sequencing
2.4. Bioinformatic Analyses
2.5. Statistical Analysis
2.5.1. Principal Component Analysis
2.5.2. Redundancy Analysis
2.5.3. Principal Coordinates Analysis
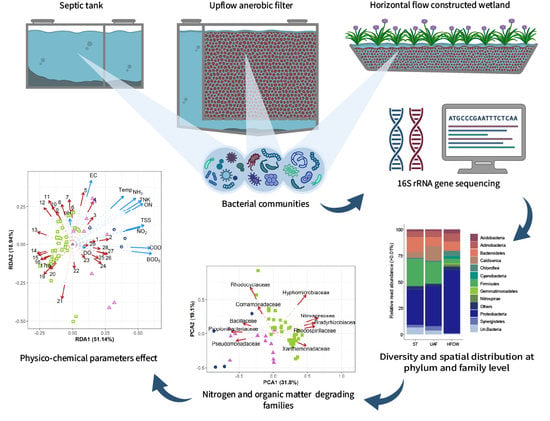
3. Results and Discussion
3.1. System Performance
3.2. Diversity and Composition of the Bacterial Communities
3.3. Spatial Variations of Nitrogen and Organic Matter Degrading Families
3.3.1. Nitrogen Degrading Bacterial Families
3.3.2. Families Degrading Organic Matter
3.4. Effect of Physicochemical Parameters on Bacterial Communities
3.5. Bacterial Communities in Multi-Stage Wwtps Located in Subtropical Regions
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Water and the Sustainable Development Goals (SDGs)|2015 UN-Water Annual International Zaragoza Conference. Water and Sustainable Development: From Vision to Action. 15–17 January 2015. Available online: https://www.un.org/waterforlifedecade/waterandsustainabledevelopment2015/open_working_group_sdg.shtml (accessed on 4 November 2020).
- Zurita, F.; Roy, E.D.; White, J.R. Municipal Wastewater Treatment in Mexico: Current Status and Opportunities for Employing Ecological Treatment Systems. Environ. Technol. 2012, 33, 1151–1158. [Google Scholar] [CrossRef]
- De Anda, J.; López-López, A.; Villegas-García, E.; Valdivia-Aviña, K. High-Strength Domestic Wastewater Treatment and Reuse with Onsite Passive Methods. Water 2018, 10, 99. [Google Scholar] [CrossRef] [Green Version]
- Fernández del Castillo, A.; Verduzco Garibay, M.; Senés-Guerrero, C.; Yebra-Montes, C.; de Anda, J.; Gradilla-Hernández, M.S. Mathematical Modeling of a Domestic Wastewater Treatment System Combining a Septic Tank, an Up Flow Anaerobic Filter, and a Constructed Wetland. Water 2020, 12, 3019. [Google Scholar] [CrossRef]
- Álvarez, J.A.; Ruíz, I.; Soto, M. Anaerobic Digesters as a Pretreatment for Constructed Wetlands. Ecol. Eng. 2008, 33, 54–67. [Google Scholar] [CrossRef]
- Zhang, L.; De Vrieze, J.; Hendrickx, T.L.G.; Wei, W.; Temmink, H.; Rijnaarts, H.; Zeeman, G. Anaerobic Treatment of Raw Domestic Wastewater in a UASB-Digester at 10 °C and Microbial Community Dynamics. Chem. Eng. J. 2018, 334, 2088–2097. [Google Scholar] [CrossRef]
- Xiong, W.; Wang, L.; Zhou, N.; Fan, A.; Wang, S.; Su, H. High-Strength Anaerobic Digestion Wastewater Treatment by Aerobic Granular Sludge in a Step-by-Step Strategy. J. Environ. Manag. 2020, 262, 110245. [Google Scholar] [CrossRef]
- Abedi, T.; Mojiri, A. Constructed Wetland Modified by Biochar/Zeolite Addition for Enhanced Wastewater Treatment. Environ. Technol. Innov. 2019, 16, 100472. [Google Scholar] [CrossRef]
- Huang, J.; Cao, C.; Liu, J.; Yan, C.; Xiao, J. The Response of Nitrogen Removal and Related Bacteria within Constructed Wetlands after Long-Term Treating Wastewater Containing Environmental Concentrations of Silver Nanoparticles. Sci. Total Environ. 2019, 667, 522–531. [Google Scholar] [CrossRef]
- Tao, W. Microbial Removal and Plant Uptake of Nitrogen in Constructed Wetlands: Mesocosm Tests on Influencing Factors. Environ. Sci. Pollut. Res. 2018, 25. [Google Scholar] [CrossRef]
- Knowles, P.; Dotro, G.; Nivala, J.; García, J. Clogging in Subsurface-Flow Treatment Wetlands: Occurrence and Contributing Factors. Ecol. Eng. 2011, 37, 99–112. [Google Scholar] [CrossRef]
- Handayani, N.I.; Yuliasni, R.; Setianingsih, N.I.; Budiarto, A. Full Scale Application of Integrated Upflow Anaerobic Filter (UAF)-Constructed Wetland (CWs) in Small Scale Batik Industry Wastewater Treatment. J. Ris. Teknol. Pencegah. Pencemaran Ind. 2020, 11, 27–35. [Google Scholar] [CrossRef]
- Ruiz, I.; Álvarez, J.A.; Díaz, M.A.; Serrano, L.; Soto, M. Municipal Wastewater Treatment in an Anaerobic Digester-Constructed Wetland System. Environ. Technol. 2008, 29, 1249–1256. [Google Scholar] [CrossRef]
- Wu, H.; Zhang, J.; Ngo, H.H.; Guo, W.; Hu, Z.; Liang, S.; Fan, J.; Liu, H. A Review on the Sustainability of Constructed Wetlands for Wastewater Treatment: Design and Operation. Bioresour. Technol. 2015, 175, 594–601. [Google Scholar] [CrossRef]
- Rajan, R.J.; Sudarsan, J.S.; Nithiyanantham, S. Microbial Population Dynamics in Constructed Wetlands: Review of Recent Advancements for Wastewater Treatment. Environ. Eng. Res. 2018, 24, 181–190. [Google Scholar] [CrossRef]
- Jia, F.; Lai, C.; Chen, L.; Zeng, G.; Huang, D.; Liu, F.; Li, X.; Luo, P.; Wu, J.; Qin, L. Spatiotemporal and Species Variations in Prokaryotic Communities Associated with Sediments from Surface-Flow Constructed Wetlands for Treating Swine Wastewater. Chemosphere 2017, 185, 1–10. [Google Scholar] [CrossRef]
- Bedoya, K.; Coltell, O.; Cabarcas, F.; Alzate, J.F. Metagenomic Assessment of the Microbial Community and Methanogenic Pathways in Biosolids from a Municipal Wastewater Treatment Plant in Medellín, Colombia. Sci. Total Environ. 2019, 648, 572–581. [Google Scholar] [CrossRef]
- Zhang, C.-B.; Wang, J.; Liu, W.-L.; Zhu, S.-X.; Ge, H.-L.; Chang, S.X.; Chang, J.; Ge, Y. Effects of Plant Diversity on Microbial Biomass and Community Metabolic Profiles in a Full-Scale Constructed Wetland. Ecol. Eng. 2010, 36, 62–68. [Google Scholar] [CrossRef]
- He, T.; Guan, W.; Luan, Z.; Xie, S. Spatiotemporal Variation of Bacterial and Archaeal Communities in a Pilot-Scale Constructed Wetland for Surface Water Treatment. Appl. Microbiol. Biotechnol. 2015, 100. [Google Scholar] [CrossRef] [PubMed]
- Cao, Q.; Wang, H.; Chen, X.; Wang, R.; Liu, J. Composition and Distribution of Microbial Communities in Natural River Wetlands and Corresponding Constructed Wetlands. Ecol. Eng. 2017, 98, 40–48. [Google Scholar] [CrossRef]
- Button, M.; Nivala, J.; Weber, K.P.; Aubron, T.; Müller, R.A. Microbial Community Metabolic Function in Subsurface Flow Constructed Wetlands of Different Designs. Ecol. Eng. 2015, 80, 162–171. [Google Scholar] [CrossRef]
- Sánchez, O. Constructed Wetlands Revisited: Microbial Diversity in the–Omics Era. Microb. Ecol. 2017, 73, 722–733. [Google Scholar] [CrossRef]
- Fu, G.; Huangshen, L.; Guo, Z.; Zhou, Q.; Wu, Z. Effect of Plant-Based Carbon Sources on Denitrifying Microorganisms in a Vertical Flow Constructed Wetland. Bioresour. Technol. 2017, 224, 214–221. [Google Scholar] [CrossRef]
- Ferrera, I.; Sánchez, O. Insights into Microbial Diversity in Wastewater Treatment Systems: How Far Have We Come? Biotechnol. Adv. 2016, 34, 790–802. [Google Scholar] [CrossRef] [Green Version]
- He, G.; Yi, F.; Zhou, S.; Lin, J. Microbial Activity and Community Structure in Two Terrace-Type Wetlands Constructed for the Treatment of Domestic Wastewater. Ecol. Eng. 2014, 67, 198–205. [Google Scholar] [CrossRef]
- Cheng, Y.; Kong, J.; Li, M. High-Throughput Sequencing Analysis of Bacterial Community Spatiotemporal Distribution in Response to Clogging in Vertical Flow Constructed Wetlands. Bioresour. Technol. 2018, 248, 104–112. [Google Scholar] [CrossRef]
- Li, B.; Chen, J.; Wu, Z.; Wu, S.; Xie, S.; Liu, Y. Seasonal and Spatial Dynamics of Denitrification Rate and Denitrifier Community in Constructed Wetland Treating Polluted River Water. Int. Biodeterior. Biodegrad. 2018, 126, 143–151. [Google Scholar] [CrossRef]
- Desta, A.F.; Assefa, F.; Leta, S.; Stomeo, F.; Wamalwa, M.; Njahira, M.; Appolinaire, D. Microbial Community Structure and Diversity in an Integrated System of Anaerobic-Aerobic Reactors and a Constructed Wetland for the Treatment of Tannery Wastewater in Modjo, Ethiopia. PLOS ONE 2014, 9, e115576. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Mhuantong, W.; Liu, S.-Y.; Pisutpaisal, N.; Wongwilaiwalin, S.; Kanokratana, P.; Wang, A.-J.; Jiang, C.-Y.; Champreda, V.; Qiu, D.-R.; et al. Tropical and Temperate Wastewater Treatment Plants Assemble Different and Diverse Microbiomes. Appl. Microbiol. Biotechnol. 2021, 105, 853–867. [Google Scholar] [CrossRef] [PubMed]
- CONAGUA Resúmenes Mensuales de Temperaturas y Lluvia. Available online: https://smn.conagua.gob.mx/es/climatologia/temperaturas-y-lluvias/resumenes-mensuales-de-temperaturas-y-lluvias (accessed on 9 November 2020).
- Eaton, A.D.; Clesceri, L.S.; Rice, E.W.; Greenberg, A.E.; Franson, M.A.H. Standard Methods for the Examination of Water and Wastewater. American Public Health Association, American Water Works Association, Water Environment Federation. Port City Baltim. 2005, 21. [Google Scholar]
- Kuczynski, J.; Stombaugh, J.; Walters, W.A.; González, A.; Caporaso, J.G.; Knight, R. Using QIIME to Analyze 16S RRNA Gene Sequences from Microbial Communities. Curr. Protoc. Bioinform. 2011, 36, 10.7. [Google Scholar] [CrossRef] [Green Version]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Caporaso, J.G. Optimizing Taxonomic Classification of Marker Gene Amplicon Sequences. PeerJ Prepr. 2018, 6, e3208v2. [Google Scholar]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An Improved Greengenes Taxonomy with Explicit Ranks for Ecological and Evolutionary Analyses of Bacteria and Archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef]
- Hall, M.; Beiko, R. 16S rRNA Gene Analysis with QIIME2: Methods and Protocols. In Methods in Molecular Biology; Humana Press: New York, NY, USA, 2018; pp. 113–129. ISBN 978-1-4939-8726-9. [Google Scholar]
- Hurlbert, S.H. The Nonconcept of Species Diversity: A Critique and Alternative Parameters. Ecology 1971, 52, 577–586. [Google Scholar] [CrossRef] [PubMed]
- Heck, K.L., Jr.; van Belle, G.; Simberloff, D. Explicit Calculation of the Rarefaction Diversity Measurement and the Determination of Sufficient Sample Size. Ecology 1975, 56, 1459–1461. [Google Scholar] [CrossRef]
- Love, M.; Anders, S.; Huber, W. Differential Analysis of Count Data–the DESeq2 Package. Genome Biol 2014, 15, 10.1186. [Google Scholar]
- Everitt, B.; Hothorn, T. An Introduction to Applied Multivariate Analysis with R; Springer Science & Business Media: Heidelberg/Berlin, Germany, 2011; ISBN 1-4419-9650-8. [Google Scholar]
- Zuur, A.; Leno, E.N.; Smith, G.M. Analyzing Ecological Data; Springer: New York, NY, USA, 2011; ISBN 978-0-387-45972-1. [Google Scholar]
- Borcard, D.; Gillet, F.; Legendre, P. Numerical Ecology with R; Springer: New York, NY, USA, 2011; ISBN 978-1-4419-7976-6. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.; O’Hara, R.; Simpson, G.; Solymos, P.; Stevenes, M.; Wagner, H. Vegan: Community Ecology Package. R Package Version 2.0-2. 2012. Available online: https://www.researchgate.net/publication/282247686_Vegan_Community_Ecology_Package_R_package_version_20-2 (accessed on 12 April 2021).
- Clarke, K.R. Non-parametric Multivariate Analyses of Changes in Community Structure. Aust. J. Ecol. 1993, 18, 117–143. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 3-319-24277-6. [Google Scholar]
- Dixon, P. VEGAN, a Package of R Functions for Community Ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Pind, P.F.; Angelidaki, I.; Ahring, B.K.; Stamatelatou, K.; Lyberatos, G. Monitoring and Control of Anaerobic Reactors. In Biomethanation II; Ahring, B.K., Ahring, B.K., Angelidaki, I., Dolfing, J., EUegaard, L., Gavala, H.N., Haagensen, F., Mogensen, A.S., Lyberatos, G., Pind, P.F., et al., Eds.; Advances in Biochemical Engineering/Biotechnology; Springer: Berlin/Heidelberg, Germany, 2003; pp. 135–182. ISBN 978-3-540-45838-8. [Google Scholar]
- Tchobanoglus, G.; Burton, F.; Stensel, H.D. Wastewater Engineering: Treatment and Reuse. Am. Water Works Assoc. J. 2003, 95, 201. [Google Scholar]
- Zhao, C.; Xie, H.; Xu, J.; Xu, X.; Zhang, J.; Hu, Z.; Liu, C.; Liang, S.; Wang, Q.; Wang, J. Bacterial Community Variation and Microbial Mechanism of Triclosan (TCS) Removal by Constructed Wetlands with Different Types of Plants. Sci. Total Environ. 2015, 505, 633–639. [Google Scholar] [CrossRef]
- Zhang, L.; Shen, Z.; Fang, W.; Gao, G. Composition of Bacterial Communities in Municipal Wastewater Treatment Plant. Sci. Total Environ. 2019, 689, 1181–1191. [Google Scholar] [CrossRef]
- Usharani, B. Metagenomics Study of the Microbes in Constructed Wetland System Treating Sewage. Int. Lett. Nat. Sci. 2019, 74, 26–48. [Google Scholar]
- Ansola, G.; Arroyo, P.; Sáenz de Miera, L.E. Characterisation of the Soil Bacterial Community Structure and Composition of Natural and Constructed Wetlands. Sci. Total Environ. 2014, 473–474, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Meng, P.; Pei, H.; Hu, W.; Shao, Y.; Li, Z. How to Increase Microbial Degradation in Constructed Wetlands: Influencing Factors and Improvement Measures. Bioresour. Technol. 2014, 157, 316–326. [Google Scholar] [CrossRef]
- Ma, S.; Ma, H.; Hu, H.; Ren, H. Effect of Mixing Intensity on Hydrolysis and Acidification of Sewage Sludge in Two-Stage Anaerobic Digestion: Characteristics of Dissolved Organic Matter and the Key Microorganisms. Water Res. 2019, 148, 359–367. [Google Scholar] [CrossRef]
- Li, X.; Li, Y.; Li, Y.; Wu, J. Diversity and Distribution of Bacteria in a Multistage Surface Flow Constructed Wetland to Treat Swine Wastewater in Sediments. Appl. Microbiol. Biotechnol. 2018, 102, 10755–10765. [Google Scholar] [CrossRef]
- Nierychlo, M.; Andersen, K.S.; Xu, Y.; Green, N.; Jiang, C.; Albertsen, M.; Dueholm, M.S.; Nielsen, P.H. MiDAS 3: An Ecosystem-Specific Reference Database, Taxonomy and Knowledge Platform for Activated Sludge and Anaerobic Digesters Reveals Species-Level Microbiome Composition of Activated Sludge. Water Res. 2020, 182, 115955. [Google Scholar] [CrossRef]
- Iguchi, A.; Terada, T.; Narihiro, T.; Yamaguchi, T.; Kamagata, Y.; Sekiguchi, Y. In Situ Detection and Quantification of Uncultured Members of the Phylum Nitrospirae Abundant in Methanogenic Wastewater Treatment Systems. Microbes Environ. 2008, advpub, 0903100063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Wen, Y.; Tang, Z.; Huang, J.; Zhou, Q.; Vymazal, J. Effects of Plant Biomass on Bacterial Community Structure in Constructed Wetlands Used for Tertiary Wastewater Treatment. Ecol. Eng. 2015, 84, 38–45. [Google Scholar] [CrossRef]
- Adrados, B.; Sánchez, O.; Arias, C.A.; Becares, E.; Garrido, L.; Mas, J.; Brix, H.; Morató, J. Microbial Communities from Different Types of Natural Wastewater Treatment Systems: Vertical and Horizontal Flow Constructed Wetlands and Biofilters. Water Res. 2014, 55, 304–312. [Google Scholar] [CrossRef] [Green Version]
- Hanada, S.; Sekiguchi, Y. The Phylum Gemmatimonadetes. In The Prokaryotes: Other Major Lineages of Bacteria and The Archaea; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 677–681. ISBN 978-3-642-38954-2. [Google Scholar]
- Vohla, C.; Alas, R.; Nurk, K.; Baatz, S.; Mander, Ü. Dynamics of Phosphorus, Nitrogen and Carbon Removal in a Horizontal Subsurface Flow Constructed Wetland. Contam. Nat. Constr. Wetl. Pollut. Dyn. Control 2007, 380, 66–74. [Google Scholar] [CrossRef]
- Xu, D.; Liu, S.; Chen, Q.; Ni, J. Microbial Community Compositions in Different Functional Zones of Carrousel Oxidation Ditch System for Domestic Wastewater Treatment. AMB Express 2017, 7, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, K.; Yue, Q.; Kong, J.; Zhao, P.; Gao, Y.; Fu, K.; Gao, B. Microbial Diversity in Combined UAF–UBAF System with Novel Sludge and Coal Cinder Ceramic Fillers for Tetracycline Wastewater Treatment. Chem. Eng. J. 2016, 285, 319–330. [Google Scholar] [CrossRef]
- Xu, S.; Yao, J.; Ainiwaer, M.; Hong, Y.; Zhang, Y. Analysis of Bacterial Community Structure of Activated Sludge from Wastewater Treatment Plants in Winter. BioMed Res. Int. 2018, 2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, S.; Wang, Y.; Li, C.; Li, Y.; Zhou, J. The Nitrogen Removal Performance and Microbial Communities in a Two-Stage Deep Sequencing Constructed Wetland for Advanced Treatment of Secondary Effluent. Bioresour. Technol. 2018, 248, 82–88. [Google Scholar] [CrossRef]
- Vymazal, J. Plants Used in Constructed Wetlands with Horizontal Subsurface Flow: A Review. Hydrobiologia 2011, 674, 133–156. [Google Scholar] [CrossRef]
- Adhikari, J.R.; Lohani, S.P. Design, Installation, Operation and Experimentation of Septic Tank–UASB Wastewater Treatment System. Renew. Energy 2019, 143, 1406–1415. [Google Scholar] [CrossRef]
- McFarlane, P.N.; Melcer, H. Pilot-Scale Evaluation of Design Criteria for Anaerobic Photosynthetic Lagoons Treating Fellmongery (Unhairing) Wastewater. Water Res. 1981, 15, 609–613. [Google Scholar] [CrossRef]
- Meyer, D.D.; de Andrade, P.A.M.; Durrer, A.; Andreote, F.D.; Corção, G.; Brandelli, A. Bacterial Communities Involved in Sulfur Transformations in Wastewater Treatment Plants. Appl. Microbiol. Biotechnol. 2016, 100, 10125–10135. [Google Scholar] [CrossRef]
- Liao, R.; Shen, K.; Li, A.-M.; Shi, P.; Li, Y.; Shi, Q.; Wang, Z. High-Nitrate Wastewater Treatment in an Expanded Granular Sludge Bed Reactor and Microbial Diversity Using 454 Pyrosequencing Analysis. Bioresour. Technol. 2013, 134, 190–197. [Google Scholar] [CrossRef]
- Wang, Q.; Hu, Y.; Xie, H.; Yang, Z. Constructed Wetlands: A Review on the Role of Radial Oxygen Loss in the Rhizosphere by Macrophytes. Water 2018, 10, 678. [Google Scholar] [CrossRef] [Green Version]
- Stottmeister, U.; Wießner, A.; Kuschk, P.; Kappelmeyer, U.; Kästner, M.; Bederski, O.; Müller, R.A.; Moormann, H. Effects of Plants and Microorganisms in Constructed Wetlands for Wastewater Treatment. Biotechnol. Adv. 2003, 22, 93–117. [Google Scholar] [CrossRef]
- Zhang, B.; Xu, X.; Zhu, L. Activated Sludge Bacterial Communities of Typical Wastewater Treatment Plants: Distinct Genera Identification and Metabolic Potential Differential Analysis. AMB Express 2018, 8, 184. [Google Scholar] [CrossRef]
- Saeed, T.; Sun, G. A Review on Nitrogen and Organics Removal Mechanisms in Subsurface Flow Constructed Wetlands: Dependency on Environmental Parameters, Operating Conditions and Supporting Media. J. Environ. Manag. 2012, 112, 429–448. [Google Scholar] [CrossRef]
- Pincam, T.; Brix, H.; Jampeetong, A. Treatment of Anaerobic Digester Effluent Using Acorus Calamus: Effects on Plant Growth and Tissue Composition. Plants 2018, 7, 36. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.-B.; Miao, M.-S.; Kong, Q.; Ni, S.-Q. Evaluation of Microbial Diversity of Activated Sludge in a Municipal Wastewater Treatment Plant of Northern China by High-Throughput Sequencing Technology. Desalination Water Treat. 2016, 57, 23516–23521. [Google Scholar] [CrossRef]
- Wu, H.; Wang, X.; He, X.; Zhang, S.; Liang, R.; Shen, J. Effects of Root Exudates on Denitrifier Gene Abundance, Community Structure and Activity in a Micro-Polluted Constructed Wetland. Sci. Total Environ. 2017, 598, 697–703. [Google Scholar] [CrossRef]
- Si, Z.; Song, X.; Wang, Y.; Cao, X.; Zhao, Y.; Wang, B.; Chen, Y.; Arefe, A. Intensified Heterotrophic Denitrification in Constructed Wetlands Using Four Solid Carbon Sources: Denitrification Efficiency and Bacterial Community Structure. Bioresour. Technol. 2018, 267, 416–425. [Google Scholar] [CrossRef]
- Heeg, K.; Pohl, M.; Sontag, M.; Mumme, J.; Klocke, M.; Nettmann, E. Microbial Communities Involved in Biogas Production from Wheat Straw as the Sole Substrate within a Two-Phase Solid-State Anaerobic Digestion. Syst. Appl. Microbiol. 2014, 37, 590–600. [Google Scholar] [CrossRef]
- Gagen, E.J.; Padmanabha, J.; Denman, S.E.; McSweeney, C.S. Hydrogenotrophic Culture Enrichment Reveals Rumen Lachnospiraceae and Ruminococcaceae Acetogens and Hydrogen-Responsive Bacteroidetes from Pasture-Fed Cattle. FEMS Microbiol. Lett. 2015, 362. [Google Scholar] [CrossRef] [Green Version]
- Abdelgadir, A.; Chen, X.; Liu, J.; Xie, X.; Zhang, J.; Zhang, K.; Wang, H.; Liu, N. Characteristics, Process Parameters, and Inner Components of Anaerobic Bioreactors. BioMed Res. Int. 2014, 2014, 841573. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.; Wang, Z.; Zou, X.; Feng, J.; Wu, Z. Microbial Communities in an Anaerobic Dynamic Membrane Bioreactor (AnDMBR) for Municipal Wastewater Treatment: Comparison of Bulk Sludge and Cake Layer. Process Biochem. 2013, 48, 510–516. [Google Scholar] [CrossRef]
- Jang, H.M.; Kim, J.H.; Ha, J.H.; Park, J.M. Bacterial and Methanogenic Archaeal Communities during the Single-Stage Anaerobic Digestion of High-Strength Food Wastewater. Bioresour. Technol. 2014, 165, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, M.L.B.; Cantão, M.E.; Mezzari, M.P.; Ma, J.; Nossa, C.W. Assessment of Bacterial and Archaeal Community Structure in Swine Wastewater Treatment Processes. Microb. Ecol. 2015, 70, 77–87. [Google Scholar] [CrossRef]
- Shu, D.; He, Y.; Yue, H.; Wang, Q. Microbial Structures and Community Functions of Anaerobic Sludge in Six Full-Scale Wastewater Treatment Plants as Revealed by 454 High-Throughput Pyrosequencing. Bioresour. Technol. 2015, 186, 163–172. [Google Scholar] [CrossRef]
- Jia, S.; Han, H.; Zhuang, H.; Hou, B. The Pollutants Removal and Bacterial Community Dynamics Relationship within a Full-Scale British Gas/Lurgi Coal Gasification Wastewater Treatment Using a Novel System. Bioresour. Technol. 2016, 200, 103–110. [Google Scholar] [CrossRef]
- Zhong, F.; Wu, J.; Dai, Y.; Yang, L.; Zhang, Z.; Cheng, S.; Zhang, Q. Bacterial Community Analysis by PCR-DGGE and 454-Pyrosequencing of Horizontal Subsurface Flow Constructed Wetlands with Front Aeration. Appl. Microbiol. Biotechnol. 2015, 99, 1499–1512. [Google Scholar] [CrossRef]
- Van Lier, J.B.; Mahmoud, N.; Zeeman, G. Anaerobic Wastewater Treatment; IWA Publishing: London, UK, 2008; pp. 415–456. [Google Scholar]
- Rajagopal, R.; Choudhury, M.R.; Anwar, N.; Goyette, B.; Rahaman, M.S. Influence of Pre-Hydrolysis on Sewage Treatment in an Up-Flow Anaerobic Sludge BLANKET (UASB) Reactor: A Review. Water 2019, 11, 372. [Google Scholar] [CrossRef] [Green Version]
- Enitan-Folami, A.; Kumari, S.; Swalaha, F.; Odiyo, J.; Bux, F. Microbiota of a Full-Scale UASB Reactor Treating Brewery Wastewater Using Illumina MiSeq Sequencing. Open Microbiol. J. 2019, 13, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Sidhu, C.; Vikram, S.; Pinnaka, A.K. Unraveling the Microbial Interactions and Metabolic Potentials in Pre- and Post-Treated Sludge from a Wastewater Treatment Plant Using Metagenomic Studies. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef]
- Kim, Y.K.; Yoo, K.; Kim, M.S.; Han, I.; Lee, M.; Kang, B.R.; Lee, T.K.; Park, J. The Capacity of Wastewater Treatment Plants Drives Bacterial Community Structure and Its Assembly. Sci. Rep. 2019, 9, 14809. [Google Scholar] [CrossRef] [Green Version]
- Krasnits, E.; Friedler, E.; Sabbah, I.; Beliavski, M.; Tarre, S.; Green, M. Spatial Distribution of Major Microbial Groups in a Well Established Constructed Wetland Treating Municipal Wastewater. Ecol. Eng. 2009, 35, 1085–1089. [Google Scholar] [CrossRef]
- Liu, L.; Li, J.; Fan, H.; Huang, X.; Wei, L.; Liu, C. Fate of Antibiotics from Swine Wastewater in Constructed Wetlands with Different Flow Configurations. Int. Biodeterior. Biodegrad. 2019, 140, 119–125. [Google Scholar] [CrossRef]
- Rizvi, H.; Ahmad, N.; Abbas, F.; Bukhari, I.H.; Yasar, A.; Ali, S.; Yasmeen, T.; Riaz, M. Start-up of UASB Reactors Treating Municipal Wastewater and Effect of Temperature/Sludge Age and Hydraulic Retention Time (HRT) on Its Performance. Arab. J. Chem. 2015, 8, 780–786. [Google Scholar] [CrossRef] [Green Version]
- Hülsen, T.; Barry, E.M.; Lu, Y.; Puyol, D.; Batstone, D.J. Low Temperature Treatment of Domestic Wastewater by Purple Phototrophic Bacteria: Performance, Activity, and Community. Water Res. 2016, 100, 537–545. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Hu, M.; Xia, Y.; Wen, X.; Ding, K. Pyrosequencing Analysis of Bacterial Diversity in 14 Wastewater Treatment Systems in China. Appl. Environ. Microbiol. 2012, 78, 7042. [Google Scholar] [CrossRef] [Green Version]
- Miura, T.; Kita, A.; Okamura, Y.; Aki, T.; Matsumura, Y.; Tajima, T.; Kato, J.; Nakashimada, Y. Effect of Salinity on Methanogenic Propionate Degradation by Acclimated Marine Sediment-Derived Culture. Appl. Biochem. Biotechnol. 2015, 177, 1541–1552. [Google Scholar] [CrossRef]
- Hou, D.; Huang, Z.; Zeng, S.; Liu, J.; Wei, D.; Deng, X.; Weng, S.; He, Z.; He, J. Environmental Factors Shape Water Microbial Community Structure and Function in Shrimp Cultural Enclosure Ecosystems. Front. Microbiol. 2017, 8, 2359. [Google Scholar] [CrossRef] [Green Version]
- Grattieri, M.; Minteer, S.D. Microbial Fuel Cells in Saline and Hypersaline Environments: Advancements, Challenges and Future Perspectives. Bioelectrochemistry 2018, 120, 127–137. [Google Scholar] [CrossRef]
- Chen, Y.-T.; Yu, N.; Sun, Z.-Y.; Gou, M.; Xia, Z.-Y.; Tang, Y.-Q.; Kida, K. Acclimation Improves Methane Production from Molasses Wastewater with High Salinity in an Upflow Anaerobic Filter Reactor: Performance and Microbial Community Dynamics. Appl. Biochem. Biotechnol. 2020, 191, 397–411. [Google Scholar] [CrossRef] [PubMed]
- Dutta, A.; Davies, C.; Ikumi, D.S. Performance of Upflow Anaerobic Sludge Blanket (UASB) Reactor and Other Anaerobic Reactor Configurations for Wastewater Treatment: A Comparative Review and Critical Updates. J. Water Supply Res. Technol. Aqua 2018, 67, 858–884. [Google Scholar] [CrossRef]
- Liang, Y.; Zhu, H.; Bañuelos, G.; Yan, B.; Zhou, Q.; Yu, X.; Cheng, X. Constructed Wetlands for Saline Wastewater Treatment: A Review. Ecol. Eng. 2017, 98, 275–285. [Google Scholar] [CrossRef]
- Lu, S.; Hu, H.; Sun, Y.; Yang, J. Effect of Carbon Source on the Denitrification in Constructed Wetlands. J. Environ. Sci. 2009, 21, 1036–1043. [Google Scholar] [CrossRef]
- Fierer, N.; Jackson, R.B. The Diversity and Biogeography of Soil Bacterial Communities. Proc. Natl. Acad. Sci. USA 2006, 103, 626. [Google Scholar] [CrossRef] [Green Version]
- Lu, L.; Xing, D.; Ren, Z.J. Microbial Community Structure Accompanied with Electricity Production in a Constructed Wetland Plant Microbial Fuel Cell. Microb. Fuel Cells 2015, 195, 115–121. [Google Scholar] [CrossRef]
- Xu, F.; Cao, F.; Kong, Q.; Zhou, L.; Yuan, Q.; Zhu, Y.; Wang, Q.; Du, Y.; Wang, Z. Electricity Production and Evolution of Microbial Community in the Constructed Wetland-Microbial Fuel Cell. Chem. Eng. J. 2018, 339, 479–486. [Google Scholar] [CrossRef]
- Cydzik-Kwiatkowska, A. Bacterial Structure of Aerobic Granules Is Determined by Aeration Mode and Nitrogen Load in the Reactor Cycle. Bioresour. Technol. 2015, 181, 312–320. [Google Scholar] [CrossRef]
- Taubert, M.; Grob, C.; Crombie, A.; Howat, A.M.; Burns, O.J.; Weber, M.; Lott, C.; Kaster, A.-K.; Vollmers, J.; Jehmlich, N.; et al. Communal Metabolism by Methylococcaceae and Methylophilaceae Is Driving Rapid Aerobic Methane Oxidation in Sediments of a Shallow Seep near Elba, Italy. Environ. Microbiol. 2019, 21, 3780–3795. [Google Scholar] [CrossRef]
- Gray, N.D.; Sherry, A.; Grant, R.J.; Rowan, A.K.; Hubert, C.R.J.; Callbeck, C.M.; Aitken, C.M.; Jones, D.M.; Adams, J.J.; Larter, S.R.; et al. The Quantitative Significance of Syntrophaceae and Syntrophic Partnerships in Methanogenic Degradation of Crude Oil Alkanes. Environ. Microbiol. 2011, 13, 2957–2975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, S.; Kuschk, P.; Brix, H.; Vymazal, J.; Dong, R. Development of Constructed Wetlands in Performance Intensifications for Wastewater Treatment: A Nitrogen and Organic Matter Targeted Review. Water Res. 2014, 57, 40–55. [Google Scholar] [CrossRef] [PubMed]
- Westgate, P.J.; Park, C. Evaluation of Proteins and Organic Nitrogen in Wastewater Treatment Effluents. Environ. Sci. Technol. 2010, 44, 5352–5357. [Google Scholar] [CrossRef] [PubMed]









| OS (n = 24) | SP1 (n = 6) | SP2 (n = 6) | SP3 (n = 6) | SP4 (n = 6) | |
|---|---|---|---|---|---|
| DO (mg/L) | 0.5 ± 0.5 | 0.5 ± 0.5 | 0.3 ± 0.1 | 0.7 ± 0.5 | 0.6 ± 0.7 |
| EC (ms/cm) | 1.5 ± 0.2 | 1.4 ± 0.3 | 1.7 ± 0.2 | 1.6 ± 0.2 | 1.5 ± 0.1 |
| pH | 7.4 ± 0.7 | 7.8 ± 0.9 | 7.2 ± 0.7 | 7.3 ± 0.7 | 7.4 ± 0.4 |
| Temperature (°C) | 20.4 ± 2.3 | 21.0 ± 1.7 | 21.1 ± 2 | 20.9 ± 2.3 | 18.5 ± 2.3 |
| A) Nitrogen Degrading Families | B) Organic Matter Degrading Families | ||||
|---|---|---|---|---|---|
| Number | Family | Number | Family | Number | Family |
| 1 | Xanthomonadaceae | 1 | Desulfobulbaceae | 19 | Methylophilaceae |
| 2 | Caulobacteraceae | 2 | Ruminococcaceae | 20 | Sphingomonadaceae |
| 3 | Comamonadaceae | 3 | Moraxellaceae | 21 | Rhodobacteraceae |
| 4 | Chromatiaceae | 4 | Porphyromonadaceae | 22 | Xanthobacteraceae |
| 5 | Mycobacteriaceae | 5 | Bacteroidaceae | 23 | Desulfomicrobiaceae |
| 6 | Microbacteriaceae | 6 | Syntrophaceae | 24 | Lachnospiraceae |
| 7 | Rhodocyclaceae | 7 | Geobacteraceae | 25 | Mogibacteriaceae |
| 8 | Pseudomonadaceae | 8 | Desulfobacteraceae | 26 | Clostridiaceae |
| 9 | Bacillaceae | 9 | Acidaminobacteraceae | 27 | Desulfovibrionaceae |
| 10 | Propionibacteriaceae | 10 | Syntrophobacteraceae | 28 | Lactobacillaceae |
| 11 | Rhizobiaceae | 11 | Hydrogenophilaceae | 29 | Enterobacteriaceae |
| 12 | Cellulomonadaceae | 12 | Methylococcaceae | ||
| 13 | Thermodesulfovibrionaceae | 13 | Acetobacteraceae | ||
| 14 | Paenibacillaceae | 14 | Sinobacteraceae | ||
| 15 | Rhodospirillaceae | 15 | Saprospiraceae | ||
| 16 | Hyphomicrobiaceae | 16 | Chitinophagaceae | ||
| 17 | Bradyrhizobiaceae | 17 | Methylocystaceae | ||
| 18 | Nitrospiraceae | 18 | Cytophagaceae | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Verduzco Garibay, M.; Fernández del Castillo, A.; Díaz Torres, O.; de Anda, J.; Yebra-Montes, C.; Senés-Guerrero, C.; Gradilla-Hernández, M.S. Characterization of the Spatial Variation of Microbial Communities in a Decentralized Subtropical Wastewater Treatment Plant Using Passive Methods. Water 2021, 13, 1157. https://doi.org/10.3390/w13091157
Verduzco Garibay M, Fernández del Castillo A, Díaz Torres O, de Anda J, Yebra-Montes C, Senés-Guerrero C, Gradilla-Hernández MS. Characterization of the Spatial Variation of Microbial Communities in a Decentralized Subtropical Wastewater Treatment Plant Using Passive Methods. Water. 2021; 13(9):1157. https://doi.org/10.3390/w13091157
Chicago/Turabian StyleVerduzco Garibay, Marycarmen, Alberto Fernández del Castillo, Osiris Díaz Torres, José de Anda, Carlos Yebra-Montes, Carolina Senés-Guerrero, and Misael Sebastián Gradilla-Hernández. 2021. "Characterization of the Spatial Variation of Microbial Communities in a Decentralized Subtropical Wastewater Treatment Plant Using Passive Methods" Water 13, no. 9: 1157. https://doi.org/10.3390/w13091157