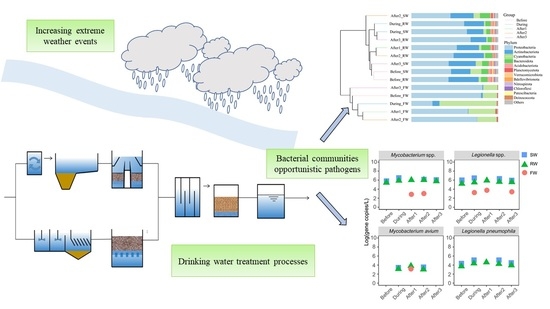
The Impact of Extreme Weather Events on Bacterial Communities and Opportunistic Pathogens in a Drinking Water Treatment Plant
Abstract
:1. Introduction
2. Materials and Methods
2.1. Water Sources
2.2. Water Treatment Process
2.3. Sample Collection
2.4. Water Chemistry Analysis
2.5. DNA Extraction and qPCR
2.6. 16S rRNA Sequencing
2.7. Statistical Analysis
3. Results
3.1. Bacterial Community Structures in Source Water and Drinking Water Treatment Plant
3.2. Changes in Bacterial Community Composition and Diversity after Extreme Weather Events
3.3. Variations in Total Bacteria and Pathogenic Microorganisms
3.4. Effect of O3-BAC Treatment on Effluent Bacterial Communities and Opportunistic Pathogens
4. Discussion
4.1. Occurrence and Removal of Total Bacteria and Opportunistic Pathogens
4.2. Impact of Extreme Weather Events on Bacteria and Opportunistic Pathogens
4.3. Contribution of BAC Tank to Downstream Bacteria and Pathogens
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Brookes, J.D.; Antenucci, J.; Hipsey, M.; Burch, M.D.; Ashbolt, N.J.; Ferguson, C. Fate and transport of pathogens in lakes and reservoirs. Environ. Int. 2004, 30, 741–759. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.; Peng, Y.; Muir, D.; Lin, J.; Zhang, X. A critical review of synthetic chemicals in surface waters of the US, the EU and China. Environ. Int. 2019, 131, 104994. [Google Scholar] [CrossRef]
- Cavicchioli, R.; Ripple, W.J.; Timmis, K.N.; Azam, F.; Bakken, L.R.; Baylis, M.; Behrenfeld, M.J.; Boetius, A.; Boyd, P.W.; Classen, A.T.; et al. Scientists’ warning to humanity: Microorganisms and climate change. Nat. Rev. Microbiol. 2019, 17, 569–586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coumou, D.; Rahmstorf, S. A decade of weather extremes. Nat. Clim. Chang. 2012, 2, 491–496. [Google Scholar] [CrossRef]
- Zhao, W. Extreme weather and climate events in China under changing climate. Nat. Sci. Rev. 2020, 7, 938–943. [Google Scholar] [CrossRef] [Green Version]
- Cann, K.F.; Thomas, D.R.; Salmon, R.L.; Wyn-Jones, A.P.; Kay, D. Extreme water-related weather events and waterborne disease. Epidemiol. Infect. 2013, 141, 671–686. [Google Scholar] [CrossRef]
- Lee, C.S.; Lee, Y.C.; Chiang, H.M. Abrupt state change of river water quality (turbidity): Effect of extreme rainfalls and typhoons. Sci. Total Environ. 2016, 557, 91–101. [Google Scholar] [CrossRef]
- Ruecker, A.; Uzun, H.; Karanfil, T.; Tsui, M.T.K.; Chow, A.T. Disinfection byproduct precursor dynamics and water treatability during an extreme flooding event in a coastal blackwater river in southeastern United States. Chemosphere 2017, 188, 90–98. [Google Scholar] [CrossRef]
- Landsman, M.R.; Rowles, L.S.; Brodfuehrer, S.H.; Maestre, J.P.; Kinney, K.A.; Kirisits, M.J.; Lawler, D.F.; Katz, L.E. Impacts of Hurricane Harvey on drinking water quality in two Texas cities. Environ. Res. Lett. 2019, 14, 124046. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.; Sevillano-Rivera, M.; Jiang, T.; Li, G.; Cotto, I.; Vosloo, S.; Carpenter, C.M.G.; Larese-Casanova, P.; Giese, R.W.; Helbling, D.E.; et al. Impact of Hurricane Maria on drinking water quality in Puerto Rico. Environ. Sci. Technol. 2020, 54, 9495–9509. [Google Scholar] [CrossRef]
- Han, Z.; An, W.; Yang, M.; Zhang, Y. Assessing the impact of source water on tap water bacterial communities in 46 drinking water supply systems in China. Water Res. 2020, 172, 115469. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Ling, F.; Zhang, M.; Liu, W.T.; Li, Y.; Liu, W. Characterization of bacterial community dynamics in a full-scale drinking water treatment plant. J. Environ. Sci. 2017, 51, 21–30. [Google Scholar] [CrossRef]
- Pinto, A.J.; Xi, C.; Raskin, L. Bacterial community structure in the drinking water microbiome is governed by filtration processes. Environ. Sci. Technol. 2012, 46, 8851–8859. [Google Scholar] [CrossRef] [PubMed]
- Kantor, R.S.; Miller, S.E.; Nelson, K.L. The water microbiome through a pilot scale advanced treatment facility for direct potable reuse. Front. Microbiol. 2019, 10, 993. [Google Scholar] [CrossRef] [Green Version]
- Proctor, C.R.; Hammes, F. Drinking water microbiology—From measurement to management. Curr. Opin. Biotechnol. 2015, 33, 87–94. [Google Scholar] [CrossRef]
- Lu, J.; Struewing, I.; Vereen, E.; Kirby, A.E.; Levy, K.; Moe, C.; Ashbolt, N. Molecular Detection of Legionella spp., their associations with Mycobacterium spp., Pseudomonas aeruginosa and amoeba hosts in a drinking water distribution system. J. Appl. Microbiol. 2016, 120, 509–521. [Google Scholar] [CrossRef]
- Li, Q.; Yu, S.; Yang, S.; Yang, W.; Que, S.; Li, W.; Qin, Y.; Yu, W.; Jiang, H.; Zhao, D. Eukaryotic community diversity and pathogenic eukaryotes in a full-scale drinking water treatment plant determined by 18S rRNA and metagenomic sequencing. Environ. Sci. Pollut. Res. 2021, 28, 17417–17430. [Google Scholar] [CrossRef]
- Ma, X.; Vikram, A.; Casson, L.; Bibby, K. Centralized drinking water treatment operations shape bacterial and Fungal community structure. Environ. Sci. Technol. 2017, 51, 7648–7657. [Google Scholar] [CrossRef]
- Hou, L.; Zhou, Q.; Wu, Q.; Gu, Q.; Sun, M.; Zhang, J. Spatiotemporal changes in bacterial community and microbial activity in a full-scale drinking water treatment plant. Sci. Total Environ. 2018, 625, 449–459. [Google Scholar] [CrossRef]
- Tiwari, A.; Hokajarvi, A.M.; Santo Domingo, J.; Elk, M.; Jayaprakash, B.; Ryu, H.; Siponen, S.; Vepsalainen, A.; Kauppinen, A.; Puurunen, O.; et al. Bacterial diversity and predicted enzymatic function in a multipurpose surface water system—From wastewater effluent discharges to drinking water production. Environ. Microbiome 2021, 16, 11. [Google Scholar] [CrossRef]
- Wang, H.; Xu, J.; Tang, W.; Li, H.; Xia, S.; Zhao, J.; Zhang, W.; Yang, Y. Removal efficacy of opportunistic pathogens and bacterial community dynamics in two drinking water treatment trains. Small 2019, 15, e1804436. [Google Scholar] [CrossRef]
- Tang, W.; Mao, Y.; Li, Q.; Meng, D.; Chen, L.; Wang, H.; Zhu, R.; Zhang, W.X. Prevalence of opportunistic pathogens and diversity of microbial communities in the water system of a pulmonary hospital. Biomed. Environ. Sci. 2020, 33, 248–259. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Edwards, M.; Falkinham, J.O.; Pruden, A. Molecular survey of the occurrence of Legionella spp., Mycobacterium spp., Pseudomonas aeruginosa, and amoeba hosts in two chloraminated drinking water distribution systems. Appl. Environ. Microbiol. 2012, 78, 6285–6294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magoc, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Gloeckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [Green Version]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq illumina sequencing platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef] [Green Version]
- Warton, D.I.; Wright, S.T.; Wang, Y. Distance-based multivariate analyses confound location and dispersion effects. Methods Ecol. Evol. 2012, 3, 89–101. [Google Scholar] [CrossRef]
- Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Sneath, P.H.A.; Sokal, R.R. Numerical Taxonomy: The Principles and Practice of Numerical Classification; Taylor & Francis, Ltd.: Oxfordshire, UK, 1975. [Google Scholar]
- Knights, D.; Kuczynski, J.; Charlson, E.S.; Zaneveld, J.; Mozer, M.C.; Collman, R.G.; Bushman, F.D.; Knight, R.; Kelley, S.T. Bayesian community-wide culture-independent microbial source tracking. Nat. Methods 2011, 8, 761–763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henry, R.; Schang, C.; Coutts, S.; Kolotelo, P.; Prosser, T.; Crosbie, N.; Grant, T.; Cottam, D.; O’Brien, P.; Deletic, A.; et al. Into the deep: Evaluation of SourceTracker for assessment of faecal contamination of coastal waters. Water Res. 2016, 93, 242–253. [Google Scholar] [CrossRef]
- Staley, C.; Kaiser, T.; Lobos, A.; Ahmed, W.; Harwood, V.J.; Brown, C.M.; Sadowsky, M.J. Application of SourceTracker for accurate identification of fecal pollution in recreational freshwater: A double-blinded study. Environ. Sci. Technol. 2018, 52, 4207–4217. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Wang, J.; Wu, Y.; Ren, C.; Song, C.; Yang, J.; Yu, H.; Giesy, J.P.; Zhang, X. Using in situ bacterial communities to monitor contaminants in river sediments. Environ. Pollut. 2016, 212, 348–357. [Google Scholar] [CrossRef]
- Ji, W.T.; Hsu, B.M.; Chang, T.Y.; Hsu, T.K.; Kao, P.M.; Huang, K.H.; Tsai, S.F.; Huang, Y.L.; Fan, C.W. Surveillance and evaluation of the infection risk of free-living amoebae and Legionella in different aquatic environments. Sci. Total Environ. 2014, 499, 212–219. [Google Scholar] [CrossRef]
- Richardson, H.; Rhodes, G.; Henrys, P.; Sedda, L.; Weightman, A.J.; Pickup, R.W. Presence of Mycobacterium avium subspecies paratuberculosis monitored over varying temporal and spatial scales in river catchments: Persistent routes for human exposure. Microorganisms 2019, 7, 136. [Google Scholar] [CrossRef] [Green Version]
- Qin, T.; Ren, H.; Chen, D.; Zhou, H.; Jiang, L.; Wu, D.; Shen, J.; Pei, F. National surveillance of legionnaires’ disease, China, 2014–2016. Emerg. Infect. Dis. 2019, 25, 1218–1219. [Google Scholar] [CrossRef]
- Yi, H.; Fang, J.; Huang, J.; Liu, B.; Qu, J.; Zhou, M. Legionella pneumophila as cause of severe community-acquired pneumonia, China. Emerg. Infect. Dis. 2020, 26, 160–162. [Google Scholar] [CrossRef]
- Dong, P.; Cui, Q.; Fang, T.; Huang, Y.; Wang, H. Occurrence of antibiotic resistance genes and bacterial pathogens in water and sediment in urban recreational water. J. Environ. Sci. 2019, 77, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Tung, M.C.; Chang, T.Y.; Hsu, B.M.; Shen, S.M.; Huang, J.T.; Kao, P.M.; Chiu, Y.C.; Fan, C.W.; Huang, Y.L. Seasonal distribution of Legionella spp., L. pneumophila in a river in Taiwan evaluated with culture-confirmed and direct DNA extraction methods. J. Hydrol. 2013, 496, 100–106. [Google Scholar] [CrossRef]
- Francy, D.S.; Brady, A.M.G.; Cicale, J.R.; Dalby, H.D.; Stelzer, E.A. Nowcasting methods for determining microbiological water quality at recreational beaches and drinking-water source waters. J. Microbiol. Meth. 2020, 175, 105970. [Google Scholar] [CrossRef] [PubMed]
- Hrudey, S.E. Chlorination disinfection by-products, public health risk tradeoffs and me. Water Res. 2009, 43, 2057–2092. [Google Scholar] [CrossRef] [PubMed]
- Ledesma, J.L.J.; Köhler, S.; Futter, M.N. Long-term dynamics of dissolved organic carbon: Implications for drinking water supply. Sci. Total Environ. 2012, 432, 1–11. [Google Scholar] [CrossRef]
- Pinto, A.J.; Schroeder, J.; Lunn, M.; Sloan, W.; Raskin, L. Spatial-temporal survey and occupancy-abundance modeling to predict bacterial community dynamics in the drinking water microbiome. mBio 2014, 5, e01135-14. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Tang, W.; Ma, J.; Wang, H. Comparison of microbial community shifts in two parallel multi-step drinking water treatment processes. Appl. Microbiol. Biotechnol. 2017, 101, 5531–5541. [Google Scholar] [CrossRef]
- Fu, Y.; Peng, H.; Liu, J.; Nguyen, T.H.; Hashmi, M.Z.; Shen, C. Occurrence and quantification of culturable and viable but non-culturable (VBNC) pathogens in biofilm on different pipes from a metropolitan drinking water distribution system. Sci. Total Environ. 2020, 764, 142851. [Google Scholar] [CrossRef] [PubMed]
- Le Dantec, C.; Duguet, J.P.; Montiel, A.; Dumoutier, N.; Dubrou, S.; Vincent, V. Chlorine disinfection of atypical mycobacteria isolated from a water distribution system. Appl. Environ. Microbiol. 2002, 68, 1025–1032. [Google Scholar] [CrossRef] [Green Version]
- Yan, D. Application of a high efficiency treating-process in the extension project of Weicun water works. Water Purif. Technol. 2010, 29, 74–78. (In Chinese) [Google Scholar] [CrossRef]
- Jiang, S.C.; Han, M.; Chandrasekaran, S.; Fang, Y.; Kellogg, C.A. Assessing the water quality impacts of two Category-5 hurricanes on St. Thomas, Virgin Islands. Water Res. 2020, 171, 115440. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Liu, J.; Chen, S.; Cheng, X.; Zheng, Z. Uniqueness of Lekima compared to tropical cyclones landed in the east coast of China during 1979–2019. Acta Oceanol. Sin. 2020, 39, 121–124. [Google Scholar] [CrossRef]
- Joh, G.; Lee, J. Cyanobacterial biofilms on sedimentation basins in a water treatment plant in South Korea. J. Appl. Phycol. 2011, 24, 285–293. [Google Scholar] [CrossRef]
- Qin, B.; Zhu, G.; Gao, G.; Zhang, Y.; Li, W.; Paerl, H.W.; Carmichael, W.W. A drinking water crisis in Lake Taihu, China: Linkage to climatic variability and lake management. Environ. Manag. 2010, 45, 105–112. [Google Scholar] [CrossRef]
- Weber, S.J.; Mishra, D.R.; Wilde, S.B.; Kramer, E. Risks for cyanobacterial harmful algal blooms due to land management and climate interactions. Sci. Total Environ. 2020, 703, 134608. [Google Scholar] [CrossRef]
- Falconer, I.R.; Humpage, A.R. Health risk assessment of cyanobacterial (blue-green algal) toxins in drinking water. Int. J. Environ. Res. Public Health 2005, 2, 43–50. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Ersan, M.S.; Wagner, E.; Plewa, M.J.; Amy, G.; Karanfil, T. Toxicity of chlorinated algal-impacted waters: Formation of disinfection byproducts vs. reduction of cyanotoxins. Water Res. 2020, 184, 116145. [Google Scholar] [CrossRef]
- Henry, R.; Harris, A.R.; Fidan, E.N.; Nelson, N.G.; Emanuel, R.E.; Jass, T.; Kathariou, S.; Niedermeyer, J.; Sharara, M.; de los Reyes, F.L.; et al. Microbial contamination in environmental waters of rural and agriculturally-dominated landscapes following Hurricane Florence. ACS ES&T Water 2021, 1, 2012–2019. [Google Scholar] [CrossRef]
- Kapoor, V.; Gupta, I.; Pasha, A.B.M.T.; Phan, D. Real-time quantitative PCR measurements of fecal indicator bacteria and human-associated source tracking markers in a Texas river following Hurricane Harvey. Environ. Sci. Technol. Lett. 2018, 5, 322–328. [Google Scholar] [CrossRef]
- Yu, P.; Zaleski, A.; Li, Q.; He, Y.; Mapili, K.; Pruden, A.; Alvarez, P.J.J.; Stadler, L.B. Elevated levels of pathogenic indicator bacteria and antibiotic resistance genes after Hurricane Harvey’s flooding in Houston. Environ. Sci. Technol. Lett. 2018, 5, 481–486. [Google Scholar] [CrossRef]
- Honda, J.R.; Virdi, R.; Chan, E.D. Global environmental nontuberculous mycobacteria and their contemporaneous man-made and natural niches. Front. Microbiol. 2018, 9, 2029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishiuchi, Y.; Iwamoto, T.; Maruyama, F. Infection sources of a common non-tuberculous mycobacterial pathogen, Mycobacterium avium complex. Front. Med. 2017, 4, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Heijnsbergen, E.; de Roda Husman, A.M.; Lodder, W.J.; Bouwknegt, M.; Docters van Leeuwen, A.E.; Bruin, J.P.; Euser, S.M.; den Boer, J.W.; Schalk, J.A.C. Viable Legionella pneumophila bacteria in natural soil and rainwater puddles. J. Appl. Microbiol. 2014, 117, 882–890. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, F.R.; Vazoller, R.F.; Foronda, A.S.; Pellizari, V.H. Phylogenetic study of Legionella species in pristine and polluted aquatic samples from a tropical Atlantic forest ecosystem. Curr. Microbiol. 2007, 55, 288–293. [Google Scholar] [CrossRef] [PubMed]
- Tryland, I.; Robertson, L.; Blankenberg, A.G.B.; Lindholm, M.; Rohrlack, T.; Liltved, H. Impact of rainfall on microbial contamination of surface water. Int. J. Clim. Chang. Strateg. Manag. 2011, 3, 361–373. [Google Scholar] [CrossRef]
- Albers, C.N.; Ellegaard-Jensen, L.; Harder, C.B.; Rosendahl, S.; Knudsen, B.E.; Ekelund, F.; Aamand, J. Groundwater chemistry determines the prokaryotic community structure of waterworks sand filters. Environ. Sci. Technol. 2015, 49, 839–846. [Google Scholar] [CrossRef]
- Tian, R.; Ning, D.; He, Z.; Zhang, P.; Spencer, S.J.; Gao, S.; Shi, W.; Wu, L.; Zhang, Y.; Yang, Y.; et al. Small and mighty: Adaptation of superphylum Patescibacteria to groundwater environment drives their genome simplicity. Microbiome 2020, 8, 51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.; Yu, S.; Li, L.; Liu, G.; Gu, Z.; Liu, M.; Liu, Z.; Ye, Y.; Xia, Q.; Ren, L. Microbial communities shaped by treatment processes in a drinking water treatment plant and their contribution and threat to drinking water safety. Front. Microbiol. 2017, 8, 2465. [Google Scholar] [CrossRef] [Green Version]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, W.; Liu, Y.; Li, Q.; Chen, L.; Li, Q.; Li, P.; Xia, S. The Impact of Extreme Weather Events on Bacterial Communities and Opportunistic Pathogens in a Drinking Water Treatment Plant. Water 2022, 14, 54. https://doi.org/10.3390/w14010054
Tang W, Liu Y, Li Q, Chen L, Li Q, Li P, Xia S. The Impact of Extreme Weather Events on Bacterial Communities and Opportunistic Pathogens in a Drinking Water Treatment Plant. Water. 2022; 14(1):54. https://doi.org/10.3390/w14010054
Chicago/Turabian StyleTang, Wei, Yunsi Liu, Qiuyan Li, Ling Chen, Qi Li, Pan Li, and Shengji Xia. 2022. "The Impact of Extreme Weather Events on Bacterial Communities and Opportunistic Pathogens in a Drinking Water Treatment Plant" Water 14, no. 1: 54. https://doi.org/10.3390/w14010054