Comparison of Two Methods for SARS-CoV-2 Detection in Wastewater: A Case Study from Sofia, Bulgaria
Abstract
:1. Introduction
2. Materials and Methods
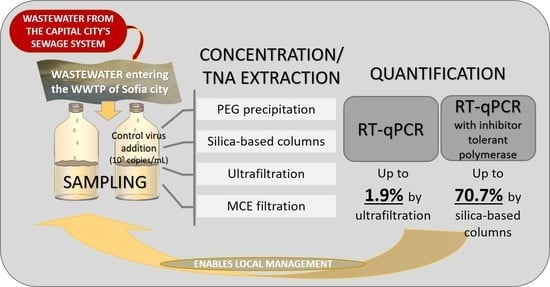
- PEG precipitation: 10% of PEG 8000 was used in combination with 2.25% NaCl for the precipitation. To sediment the viral RNA in the pellet, the mixture was centrifuged in a swinging-bucket rotor at 12,000 g for 120 min at 4 °C without braking force. The pellet was resuspended in 200 µL of nuclease-free water.
- MCE filtration: In the concentration approach based on electronegative filtration, MCE filters (Merck Millipore Ltd., Ireland, Carrigtohill) were used. Before the procedure, the pH of the samples was measured and adjusted to neutral if needed. MgCl2 was added to the sample to a final concentration of 25 mM. 50 mL of the sample was filtered through an MCE filter with a pore size of 0.45 µm using a sterile filtration funnel. The filter was then cut into pieces and immediately placed in a 2 mL bead beating tube (Zymo Research, Irvine, CA, USA) with 200 µL of saline solution (0.9% NaCl). After that, the samples were processed according to the procedure described by Ahmed (2020) [17].
- Ultrafiltration: Amicon® Ultra-15 devices with a molecular weight cutoff of 30 kDa (Merck Millipore Ltd., Ireland, Carrigtohill) were used to concentrate the samples by ultrafiltration. A volume of 50 mL was filtered through the devices by centrifugation at 4750 g for 10 min at 4 °C. Finally, a concentrate with a volume of about 200 µL was obtained.
- RT-qPCR with polymerase without inhibitor resistance—the gb Sarbeco N (primary test) kit (Generi Biotech, Czech Republic, Hradec Králové) was used according to the protocol provided by the manufacturer. The reaction volume was 20 µL, and 5 µL of the extracted TNAs were used as a matrix. To remove the effects of potential inhibitors, some samples were diluted 2- or 5-fold, and others were treated with the OneStep PCR Inhibitor Removal Kit (Zymo Research, Irvine, CA, USA). Viral levels were quantified using a standard curve prepared from falling tenfold dilutions of IDT 2019-nCoV N Positive Control Standards (IDT, USA, IA, Coralville) included in each PCR run.
- RT-qPCR with inhibitor-resistant polymerase—the Wastewater SARS-CoV-2 RT-qPCR Systems kit (Promega, Germany) targeting the N1 gene of SARS-CoV-2 was used. Viral levels were quantified by a standard curve prepared by using a series of tenfold dilutions of the standards included in the kit.
3. Results
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Safford, H.R.; Shapiro, K.; Bischel, H.N. Wastewater Analysis Can Be a Powerful Public Health Tool—If It’s Done Sensibly. Proc. Natl. Acad. Sci. USA 2022, 119, e2119600119. [Google Scholar] [CrossRef] [PubMed]
- Medema, G.; Heijnen, L.; Elsinga, G.; Italiaander, R.; Brouwer, A. Presence of SARS-Coronavirus-2 RNA in Sewage and Correlation with Reported COVID-19 Prevalence in the Early Stage of the Epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020, 7, 511–516. [Google Scholar] [CrossRef]
- Randazzo, W.; Truchado, P.; Cuevas-Ferrando, E.; Simón, P.; Allende, A.; Sánchez, G. SARS-CoV-2 RNA in Wastewater Anticipated COVID-19 Occurrence in a Low Prevalence Area. Water Res. 2020, 181, 115942. [Google Scholar] [CrossRef] [PubMed]
- Sherchan, S.P.; Shahin, S.; Ward, L.M.; Tandukar, S.; Aw, T.G.; Schmitz, B.; Ahmed, W.; Kitajima, M. First Detection of SARS-CoV-2 RNA in Wastewater in North America: A Study in Louisiana, USA. Sci. Total Environ. 2020, 743, 140621. [Google Scholar] [CrossRef]
- Zhu, Y.; Oishi, W.; Saito, M.; Kitajima, M.; Sano, D. Early Warning of COVID-19 in Tokyo via Wastewater-Based Epidemiology: How Feasible It Really Is? J. Water Environ. Technol. 2021, 19, 170–183. [Google Scholar] [CrossRef]
- Sanjuán, R.; Domingo-Calap, P. Reliability of Wastewater Analysis for Monitoring COVID-19 Incidence Revealed by a Long-Term Follow-Up Study. Front. Virol. 2021, 1, 776998. [Google Scholar] [CrossRef]
- Daughton, C.G. Wastewater Surveillance for Population-Wide Covid-19: The Present and Future. Sci. Total Environ. 2020, 736, 139631. [Google Scholar] [CrossRef]
- Sharara Id, N.; Id, N.E.; Duvallet Id, C.; Ghaeli, N.; Matus, M.; Heussner, J.; Olesenid, S.W.; Alm, E.J.; Chaiid, P.R.; Ericksonid, T.B. Wastewater Network Infrastructure in Public Health: Applications and Learnings from the COVID-19 Pandemic. PLOS Glob. Public Health 2021, 1, e0000061. [Google Scholar] [CrossRef]
- Barceló, D. Wastewater-Based Epidemiology to Monitor COVID-19 Outbreak: Present and Future Diagnostic Methods to Be in Your Radar. Case Stud. Chem. Environ. Eng. 2020, 2, 100042. [Google Scholar] [CrossRef]
- Feng, S.; Roguet, A.; McClary-Gutierrez, J.S.; Newton, R.J.; Kloczko, N.; Meiman, J.G.; McLellan, S.L. Evaluation of Sampling, Analysis, and Normalization Methods for SARS-CoV-2 Concentrations in Wastewater to Assess COVID-19 Burdens in Wisconsin Communities. ACS ES&T Water 2021, 1, 1955–1965. [Google Scholar] [CrossRef]
- Pulicharla, R.; Kaur, G.; Brar, S.K. A Year into the COVID-19 Pandemic: Rethinking of Wastewater Monitoring as a Preemptive Approach. J. Environ. Chem. Eng. 2021, 9, 106063. [Google Scholar] [CrossRef]
- Rusiñol, M.; Martínez-Puchol, S.; Forés, E.; Itarte, M.; Girones, R.; Bofill-Mas, S. Concentration Methods for the Quantification of Coronavirus and Other Potentially Pandemic Enveloped Virus from Wastewater. Curr. Opin. Environ. Sci. Health 2020, 17, 21–28. [Google Scholar] [CrossRef]
- Dimitrakopoulos, L.; Kontou, A.; Strati, A.; Galani, A.; Kostakis, M.; Kapes, V.; Lianidou, E.; Thomaidis, N.; Markou, A. Evaluation of Viral Concentration and Extraction Methods for SARS-CoV-2 Recovery from Wastewater Using Droplet Digital and Quantitative RT-PCR. Case Stud. Chem. Environ. Eng. 2022, 6, 100224. [Google Scholar] [CrossRef]
- Pérez-Cataluña, A.; Cuevas-Ferrando, E.; Randazzo, W.; Falcó, I.; Allende, A.; Sánchez, G. Comparing Analytical Methods to Detect SARS-CoV-2 in Wastewater. Sci. Total Environ. 2021, 758, 143870. [Google Scholar] [CrossRef]
- Zheng, X.; Deng, Y.; Xu, X.; Li, S.; Zhang, Y.; Ding, J.; On, H.Y.; Lai, J.C.C.; In Yau, C.; Chin, A.W.H.; et al. Comparison of Virus Concentration Methods and RNA Extraction Methods for SARS-CoV-2 Wastewater Surveillance. Sci. Total Environ. 2022, 824, 153687. [Google Scholar] [CrossRef]
- Philo, S.E.; Ong, A.Q.W.; Keim, E.K.; Swanstrom, R.; Kossik, A.L.; Zhou, N.A.; Beck, N.K.; Meschke, J.S. Development and Validation of the Skimmed Milk Pellet Extraction Protocol for SARS-CoV-2 Wastewater Surveillance. Food Environ. Virol. 2022, 1, 1–9. [Google Scholar] [CrossRef]
- Ahmed, W.; Bertsch, P.M.; Bivins, A.; Bibby, K.; Farkas, K.; Gathercole, A.; Haramoto, E.; Gyawali, P.; Korajkic, A.; McMinn, B.R.; et al. Comparison of Virus Concentration Methods for the RT-QPCR-Based Recovery of Murine Hepatitis Virus, a Surrogate for SARS-CoV-2 from Untreated Wastewater. Sci. Total Environ. 2020, 739, 139960. [Google Scholar] [CrossRef]
- Sangkham, S. A Review on Detection of SARS-CoV-2 RNA in Wastewater in Light of the Current Knowledge of Treatment Process for Removal of Viral Fragments. J. Environ. Manag. 2021, 299, 113563. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.W.; Li, J.S.; Guo, T.K.; Zhen, B.; Kong, Q.X.; Yi, B.; Li, Z.; Song, N.; Jin, M.; Xiao, W.J.; et al. Concentration and Detection of SARS Coronavirus in Sewage from Xiao Tang Shan Hospital and the 309th Hospital. J. Virol. Methods 2005, 128, 156–161. [Google Scholar] [CrossRef]
- Peinado, B.; Martínez-García, L.; Martínez, F.; Nozal, L.; Sánchez, M.B. Improved Methods for the Detection and Quantification of SARS-CoV-2 RNA in Wastewater. Sci. Rep. 2022, 12, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, M.S.; Machado, B.A.S.; Rolo, C.d.A.; Hodel, K.V.S.; Almeida, E.d.S.; de Andrade, J.B. Evaluation of SARS-CoV-2 Concentrations in Wastewater and River Water Samples. Case Stud. Chem. Environ. Eng. 2022, 6, 100214. [Google Scholar] [CrossRef]
- Juel, M.A.I.; Stark, N.; Nicolosi, B.; Lontai, J.; Lambirth, K.; Schlueter, J.; Gibas, C.; Munir, M. Performance Evaluation of Virus Concentration Methods for Implementing SARS-CoV-2 Wastewater Based Epidemiology Emphasizing Quick Data Turnaround. Sci. Total Environ. 2021, 801, 149656. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, R.; Curtis, K.; Bivins, A.; Bibby, K.; Weir, M.H.; Yetka, K.; Thompson, H.; Keeling, D.; Mitchell, J.; Gonzalez, D. COVID-19 Surveillance in Southeastern Virginia Using Wastewater-Based Epidemiology. Water Res. 2020, 186, 116296. [Google Scholar] [CrossRef] [PubMed]
- Michael-Kordatou, I.; Karaolia, P.; Fatta-Kassinos, D. Sewage Analysis as a Tool for the COVID-19 Pandemic Response and Management: The Urgent Need for Optimised Protocols for SARS-CoV-2 Detection and Quantification. J. Environ. Chem. Eng. 2020, 8, 104306. [Google Scholar] [CrossRef]
- Pecson, B.M.; Darby, E.; Haas, C.N.; Amha, Y.M.; Bartolo, M.; Danielson, R.; Dearborn, Y.; di Giovanni, G.; Ferguson, C.; Fevig, S.; et al. Sars-Cov-2 Interlaboratory Consortium. Reproducibility and Sensitivity of 36 Methods to Quantify the SARS-CoV-2 Genetic Signal in Raw Wastewater: Findings from an Interlaboratory Methods Evaluation in the U.S. Environ. Sci. 2021, 7, 504–520. [Google Scholar] [CrossRef]
- Mondal, S.; Feirer, N.; Brockman, M.; Preston, M.A.; Teter, S.J.; Ma, D.; Goueli, S.A.; Moorji, S.; Saul, B.; Cali, J.J. A Direct Capture Method for Purification and Detection of Viral Nucleic Acid Enables Epidemiological Surveillance of SARS-CoV-2. Sci. Total Environ. 2021, 795, 148834. [Google Scholar] [CrossRef]
- Kabdaşlı, I.; Tünay, O. Concentration Techniques Tailored for the Detection of SARS-CoV-2 Genetic Material in Domestic Wastewater and Treatment Plant Sludge: A Review. J. Environ. Chem. Eng. 2021, 9, 106296. [Google Scholar] [CrossRef]
- Giraud-Billoud, M.; Cuervo, P.; Altamirano, J.C.; Pizarro, M.; Aranibar, J.N.; Catapano, A.; Cuello, H.; Masachessi, G.; Vega, I.A. Monitoring of SARS-CoV-2 RNA in Wastewater as an Epidemiological Surveillance Tool in Mendoza, Argentina. Sci. Total Environ. 2021, 796, 148887. [Google Scholar] [CrossRef]
- Ahmed, W.; Simpson, S.L.; Bertsch, P.M.; Bibby, K.; Bivins, A.; Blackall, L.L.; Bofill-Mas, S.; Bosch, A.; Brandão, J.; Choi, P.M.; et al. Minimizing Errors in RT-PCR Detection and Quantification of SARS-CoV-2 RNA for Wastewater Surveillance. Sci. Total Environ. 2022, 805, 149877. [Google Scholar] [CrossRef]
- Kumblathan, T.; Piroddi, N.; Hrudey, S.E.; Li, X.F. Wastewater Based Surveillance of SARS-CoV-2: Challenges and Perspective from a Canadian Inter-Laboratory Study. J. Environ. Sci. 2022, 116, 229. [Google Scholar] [CrossRef]
- Wu, F.; Zhang, J.; Xiao, A.; Gu, X.; Lin Lee, W.; Armas, F.; Kauffman, K.; Hanage, W.; Matus, M.; Ghaeli, N.; et al. SARS-CoV-2 Titers in Wastewater Are Higher than Expected from Clinically Confirmed Cases. mSystems 2020, 5, e00614-20. [Google Scholar] [CrossRef]
- Whitney, O. Direct Wastewater RNA Capture and Purification via the “Sewage, Salt, Silica and SARS-CoV-2 (4S)” Method; protocols.io. Available online: https://doi.org/10.17504/protocols.io.biwekfbe (accessed on 1 April 2022). [CrossRef]
- Google Maps. Available online: https://maps.google.com/ (accessed on 20 December 2022).
- Srinivas, T. Environmental Biotechnology. New Age International Pvt Ltd. Publishers: New Delhi, India, 2008. [Google Scholar]
- Blanco, A.; Abid, I.; Al-Otaibi, N.; Pérez-Rodríguez, F.J.; Fuentes, C.; Guix, S.; Pintó, R.M.; Bosch, A. Glass Wool Concentration Optimization for the Detection of Enveloped and Non-Enveloped Waterborne Viruses. Food Environ. Virol. 2019, 11, 184–192. [Google Scholar] [CrossRef]
- Ye, Y.; Ellenberg, R.M.; Graham, K.E.; Wigginton, K.R. Survivability, Partitioning, and Recovery of Enveloped Viruses in Untreated Municipal Wastewater. Environ. Sci. Technol. 2016, 50, 10. [Google Scholar] [CrossRef]
- Pellegrinelli, L.; Castiglioni, S.; Cocuzza, C.E.; Bertasi, B.; Primache, V.; Schiarea, S.; Salmoiraghi, G.; Franzetti, A.; Musemeci, R.; Tilola, M.; et al. Evaluation of Pre-Analytical and Analytical Methods for Detecting SARS-CoV-2 in Municipal Wastewater Samples in Northern Italy. Water 2022, 14, 833. [Google Scholar] [CrossRef]
- Sapula, S.A.; Whittall, J.J.; Pandopulos, A.J.; Gerber, C.; Venter, H. An Optimized and Robust PEG Precipitation Method for Detection of SARS-CoV-2 in Wastewater. Sci. Total Environ. 2021, 785, 147270. [Google Scholar] [CrossRef]
- Qiu, Y.; Yu, J.; Pabbaraju, K.; Lee, B.E.; Gao, T.; Ashbolt, N.J.; Hrudey, S.E.; Diggle, M.; Tipples, G.; Maal-Bared, R.; et al. Validating and Optimizing the Method for Molecular Detection and Quantification of SARS-CoV-2 in Wastewater. Sci. Total Environ. 2022, 812, 151434. [Google Scholar] [CrossRef]
- Ahmed, W.; Smith, W.J.M.; Metcalfe, S.; Jackson, G.; Choi, P.M.; Morrison, M.; Field, D.; Gyawali, P.; Bivins, A.; Bibby, K.; et al. Comparison of RT-qPCR and RT-dPCR Platforms for the Trace Detection of SARS-CoV-2 RNA in Wastewater. ACS Environ. Sci. Technol. Water 2022. [Google Scholar] [CrossRef]
- Mazumder, P.; Dash, S.; Honda, R.; Sonne, C.; Kumar, M. Sewage Surveillance for SARS-CoV-2: Molecular Detection, Quantification, and Normalization Factors. Curr. Opin. Environ. Sci. Health 2022, 28, 100363. [Google Scholar] [CrossRef]
- Alhama, J.; Maestre, J.P.; Martín, M.Á.; Michán, C. Monitoring COVID-19 through SARS-CoV-2 Quantification in Wastewater: Progress, Challenges and Prospects. Microb. Biotechnol. 2022, 15, 1719–1728. [Google Scholar] [CrossRef]
- Philo, S.E.; Keim, E.K.; Swanstrom, R.; Ong, A.Q.W.; Burnor, E.A.; Kossik, A.L.; Harrison, J.C.; Demeke, B.A.; Zhou, N.A.; Beck, N.K.; et al. A Comparison of SARS-CoV-2 Wastewater Concentration Methods for Environmental Surveillance. Sci. Total Environ. 2021, 760, 144215. [Google Scholar] [CrossRef]
- LaTurner, Z.W.; Zong, D.M.; Kalvapalle, P.; Gamas, K.R.; Terwilliger, A.; Crosby, T.; Ali, P.; Avadhanula, V.; Santos, H.H.; Weesner, K.; et al. Evaluating Recovery, Cost, and Throughput of Different Concentration Methods for SARS-CoV-2 Wastewater-Based Epidemiology. Water Res. 2021, 197, 117043. [Google Scholar] [CrossRef] [PubMed]
- Dumke, R.; Barron, M.d.l.C.; Oertel, R.; Helm, B.; Kallies, R.; Berendonk, T.U.; Dalpke, A. Evaluation of Two Methods to Concentrate SARS-CoV-2 from Untreatedwastewater. Pathogens 2021, 10, 195. [Google Scholar] [CrossRef] [PubMed]
- Whitney, O.N.; Kennedy, L.C.; Fan, V.B.; Hinkle, A.; Kantor, R.; Greenwald, H.; Crits-Christoph, A.; Al-Shayeb, B.; Chaplin, M.; Maurer, A.C. Sewage, Salt, Silica, and SARS-CoV-2 (4S): An Economical Kit-Free Method for Direct Capture of SARS-CoV-2 RNA from Wastewater. Environ. Sci. Technol. 2021, 55, 4880–4888. [Google Scholar] [CrossRef] [PubMed]
- Flood, M.T.; D’Souza, N.; Rose, J.B.; Aw, T.G. Methods Evaluation for Rapid Concentration and Quantification of SARS-CoV-2 in Raw Wastewater Using Droplet Digital and Quantitative RT-PCR. Food Environ. Virol. 2021, 13, 303–315. [Google Scholar] [CrossRef] [PubMed]


| PEG * Concentration | Silica-Based Columns | Ultrafiltration | MCE Filtration | ||
|---|---|---|---|---|---|
| qPCR with inhibitor tolerant polymerase | Initial volume of the sample, mL | 100 mL | 40 mL | 50 mL | 50 mL |
| Need for a separate TNA extraction kit | yes | no | yes | yes | |
| Efficiency of recovery, % | 22.46% | 70.66% | 14.69% | 9.48% | |
| qPCR with conventional polymerase | Initial volume of the sample, mL | 100 mL | 50 mL | 50 mL | 50 mL |
| Need for a separate TNA extraction kit | yes | no | yes | yes | |
| Efficiency of recovery, % | 0.19% | 1.02% | 1.88% | 0.06% |
| Concentration Method | Virus | Type of Matrix | Initial Volume of the Sample, mL | Efficiency of Virus Recovery, % | Reference |
|---|---|---|---|---|---|
| PEG precipitation | Severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2) | Wastewater from municipal WWTP | 105 mL | 22.46% | This study * |
| Severe acute respiratory syndrome Coronavirus (SARSCoV) | Hospital wastewater | 100 mL | 1.02% | Wang, 2005 [19] | |
| Transmissible gastroenteritis virus (Coronaviridae) | Cell culture | Artificial wastewater with 1.78 × 106 TCID50/L | 51% | Blanco, 2019 [35] | |
| Murine hepatitis virus | Wastewater from municipal WWTP | 250 mL | ~6% | Ye, 2016 [36] | |
| Murine hepatitis virus | Wastewater from municipal WWTP | 50 mL | 44.0% | Ahmed, 2020 [17] | |
| Severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2) | Wastewater from municipal WWTP | 80–250 mL | 1%–76% | Pellegrinelli, 2022 [37] | |
| Severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2) | Wastewater from municipal WWTP | 100 mL | 27.5%–56.7% | Sapula, 2021 [38] | |
| Ultrafiltration | Severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2) | Wastewater from municipal WWTP | 50 ml | 14.69% | This study * |
| Murine hepatitis virus | Wastewater from municipal WWTP | 50 mL | 56.0% | Ahmed, 2020 [17] | |
| Mengovirus, added as a surrogate for SARS-CoV-2 | Wastewater from municipal WWTP | 100 mL | 5.74–17.59% | Peinado, 2022 [20] | |
| Human coronavirus 229E | Wastewater from municipal WWTP | 100 mL | 0.02–73.0% | Qiu, 2022 [39] | |
| Severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2) | Wastewater from municipal WWTP | 40 mL | 46.2–63.8% | Fonseca, 2022 [21] | |
| Electronegative filters | Severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2) | Wastewater from municipal WWTP | 50 mL | 9.48% | This study * |
| Murine hepatitis virus | Wastewater from municipal WWTP | 50 mL | Up to 65.7% | Ahmed, 2020 [17] | |
| Bovine coronavirus | Wastewater from municipal WWTP | 100 mL | 4.8% | Gonzalez, 2020 [23] | |
| Severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2) | Wastewater from municipal WWTP | 40 mL | 21.1–34.5% | Fonseca, 2022 [21] | |
| Silica-based columns | Severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2) | Wastewater from municipal WWTP | 40 ml | 70.66% | This study * |
| Severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2) | Wastewater from municipal WWTP | 50 mL | 38–39% | Dimitrakopoulos, 2022 [13] |
| Concentration Method | Strengths | Weaknesses | Opportunities | Threats |
|---|---|---|---|---|
| PEG precipitation | - many virological studies have applied this method, which allows for good comparability of results - low price - requires only basic equipment - the necessary components are easily accessible | - more time-consuming - laborious - requires the use of a separate kit for nucleic acid extraction, which prolongs, complicates, and makes testing more expensive concentration not only of nucleic acids but also of inhibitors in the samples | - possibility to change the starting volume of the samples - possibility of flexible use of the concentrate (adding steps for additional purification of the inhibitors) | - risk of errors due to a longer protocol - lower reproducibility of results |
| Ultrafiltration | - simple method - quick to execute - only basic equipment is needed - does not require the use of specialized chemicals - reproducible | - high price of consumables - concentration of some inhibitors together with nucleic acids - limited volume of processed samples requires the use of a separate kit for the extraction of nucleic acids, which prolongs, complicates, and makes the testing more expensive | - possibility of washing the concentrate in order to decrease the concentration of lower molecular weight inhibitors; - possibility of using membranes with a different molecular range, optimized for specific wastewaters | - high risk of shortage of specific ultrafiltration devices due to difficulties in purchaseand supply |
| Electronegative filters | - simple to implement - inexpensive - materials are routine for laboratories working with wastewater | - low efficiency of viral RNA recovery - requires the use of a separate kit for nucleic acids extraction, which prolongs, complicates, and makes the testing more expensive - substantial influence of inhibitors. | - concentration can be done from significantly larger volumes of the sample | - risk of low reproducibility due to the many steps that are not standardized and depend on the investigator - risk of filter clogging; |
| Silica-based columns | - fast method - concentration and extraction of viral RNA are combined in one protocol—no separate kit for TNA extraction is required - high efficiency - high reproducibility of the results | - a method that requires specific equipment - higher price of consumables | - possibility of automation - ready-made kits are available on the market, which makes the work even easier | - risk of supply shortages for the specific materials (columns, etc.) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Belouhova, M.; Peykov, S.; Stefanova, V.; Topalova, Y. Comparison of Two Methods for SARS-CoV-2 Detection in Wastewater: A Case Study from Sofia, Bulgaria. Water 2023, 15, 658. https://doi.org/10.3390/w15040658
Belouhova M, Peykov S, Stefanova V, Topalova Y. Comparison of Two Methods for SARS-CoV-2 Detection in Wastewater: A Case Study from Sofia, Bulgaria. Water. 2023; 15(4):658. https://doi.org/10.3390/w15040658
Chicago/Turabian StyleBelouhova, Mihaela, Slavil Peykov, Vesela Stefanova, and Yana Topalova. 2023. "Comparison of Two Methods for SARS-CoV-2 Detection in Wastewater: A Case Study from Sofia, Bulgaria" Water 15, no. 4: 658. https://doi.org/10.3390/w15040658